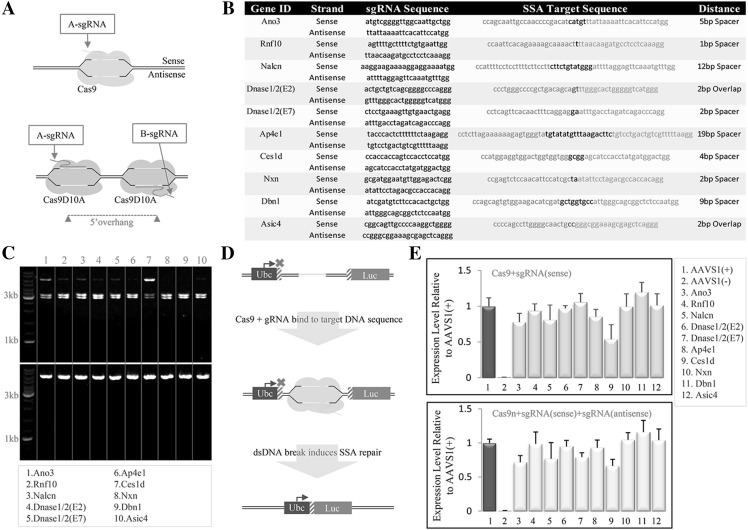
Fig. 1.
Strategy for CRISPR/Cas9(n) mediated KOMP construct targeting. a Diagram illustrating sgRNA-design strategy for use with Cas9 and/or Cas9n. The top illustration describes a single sgRNA:Cas9—complex strategy for inducing dsDNA cleavage. The bottom illustration describes a scenario in which a pair of sgRNA:Cas9n complexes are combined to induce nicks on opposite DNA strands, creating 5′ overhangs if the paired sgRNA:Cas9n complexes are not overlapping (the overlapping complexes form 3′ overhangs). Green, sgRNAs. b Table listing the sgRNAs selected for each gene in (a) (first column), the strand to which each sgRNA was designed to bind (second column), sgRNA sequences (third column), sgRNA-target sequences (blue, sgRNA target on the sense strand; red, sgRNA target on the antisense strand; black, spacer sequence between the two targets) (forth column), and spacer distance (last column). c Agarose analysis showing result of in vitro assay with Cas9 protein. d Schematic diagram illustrating the mechanism of the SSA assay. e Bar graph showing the SSA assay result relative to the AAVS1 sgRNA. Top graph, result using Cas9; bottom graph, result using Cas9n. AAVS1 (−) indicates the negative control, which is identical to AAVS1 (+) without sgRNA. (Color figure online)