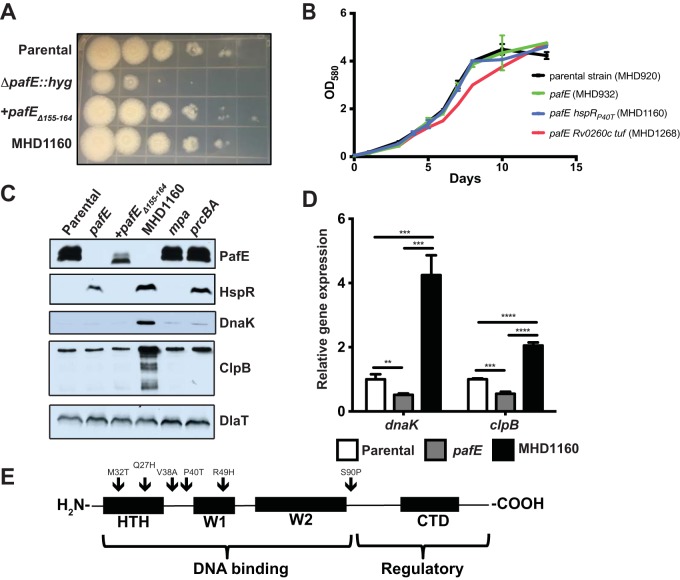
FIG 1.
Mutations in hspR rescue the growth defect of an M. tuberculosis pafE mutant. (A) The indicated M. tuberculosis strains were grown to logarithmic phase, diluted, and inoculated onto Middlebrook 7H11 agar for 2 weeks. Tenfold serial dilutions were made and spotted on agar from left (undiluted) to right (100,000-fold dilution). (B) The indicated M. tuberculosis strains were grown in 7H9-ADN, with OD580 recorded at the indicated time points. Error bars represent standard deviations for three replicates. (C) HspR targets accumulate in a pafE suppressor strain. Total cell lysates were prepared from the indicated M. tuberculosis strains. For PafE, HspR, and DlaT, samples were separated by 15% SDS-PAGE and transferred to a nitrocellulose membrane that was cut into three pieces, which were analyzed by immunoblotting with the indicated antibodies. For DnaK and ClpB, the same samples were diluted 1:20, separated by 10% SDS-PAGE, and analyzed by immunoblotting with the indicated antibodies. (D) HspR target genes are derepressed in a pafE suppressor strain. The indicated M. tuberculosis strains were grown to an OD580 of ∼0.7, and RNA was purified and reverse-transcribed into cDNA for quantitative real-time PCR analysis. Gene expression was normalized to dlaT. Statistical analysis was done by nonparametric Student's t test. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Error bars represent standard deviations for three replicates. (E) pafE suppressor mutants have mutations in hspR. pafE suppressor mutants were isolated, and the hspR gene from each was PCR amplified and sequenced. Locations of mutations are indicated by arrows on a map showing the major functional domains of HspR. HTH, helix-turn-helix domain; W, wing domain; CTD, C-terminal domain.