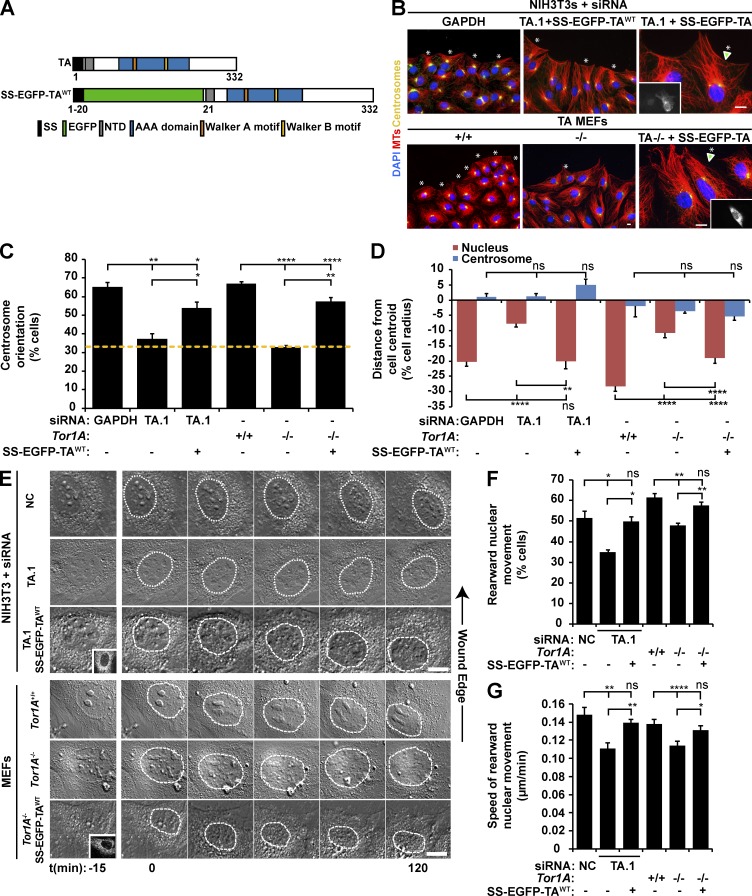
Figure 1.
TA depletion inhibits rearward nuclear movement during centrosome orientation. (A) Diagrams of the human TA protein and SS-EGFP-TAWT. Protein domains were identified using the SMART platform (Schultz et al., 1998). NTD, N-terminal domain (Vander Heyden et al., 2011). (B) Representative epifluorescence images of centrosome orientation in NIH3T3 fibroblasts treated with GAPDH or TA.1 siRNA as well as Tor1A+/+ and Tor1A−/− MEFs. TA-depleted NIH3T3 fibroblasts and Tor1A−/− MEFs expressing SS-EGFP-TAWT are also shown (arrowheads and insets). Asterisks indicate oriented centrosomes. (C) Centrosome orientation in the cells described in B. The dashed yellow line denotes random orientation, which is ∼33% (Palazzo et al., 2001). (D) Mean centrosome and nucleus positions from the cells described in B. The cell center is defined as 0. Positive values are toward the leading edge, and negative values are away. n ≥ 251. (E) Representative montages of DIC images of nuclear movement during centrosome orientation in wound-edge NIH3T3 fibroblasts treated with NC or TA.1 siRNA as well as Tor1A+/+ and Tor1A−/− MEFs. A DIC montage of wound-edge cells expressing SS-EGFP-TAWT (insets) is also provided. Nuclei are outlined by dashed white lines. Time is relative to the addition of LPA (0 min). (F) Percentages of the cells described in E with rearward moving nuclei. (G) Mean velocities of nuclear movement from cells described in E. n ≥ 14. Bars, 10 µm. Error bars show SEM. *, P < 0.05; **, P < 0.01; ****, P < 0.0001; ns, not significant.