Reduced PTEN protein is linked to tumorigenesis. Here, Chen et al. show that the nuclear transport receptor Importin-11 separates PTEN from degradation machinery. IPO11 mutant mice exhibit PTEN degradation, lung adenocarcinoma, and prostate neoplasia. In human prostatectomy patients, IPO11 status predicts disease recurrence and metastasis.
Abstract
Phosphatase and tensin homologue (PTEN) protein levels are critical for tumor suppression. However, the search for a recurrent cancer-associated gene alteration that causes PTEN degradation has remained futile. In this study, we show that Importin-11 (Ipo11) is a transport receptor for PTEN that is required to physically separate PTEN from elements of the PTEN degradation machinery. Mechanistically, we find that the E2 ubiquitin-conjugating enzyme and IPO11 cargo, UBE2E1, is a limiting factor for PTEN degradation. Using in vitro and in vivo gene-targeting methods, we show that Ipo11 loss results in degradation of Pten, lung adenocarcinoma, and neoplasia in mouse prostate with aberrantly high levels of Ube2e1 in the cytoplasm. These findings explain the correlation between loss of IPO11 and PTEN protein in human lung tumors. Furthermore, we find that IPO11 status predicts disease recurrence and progression to metastasis in patients choosing radical prostatectomy. Thus, our data introduce the IPO11 gene as a tumor-suppressor locus, which is of special importance in cancers that still retain at least one intact PTEN allele.
Introduction
The phosphoinositide 3-kinase (PI3K) pathway is among the most frequently altered growth-control mechanisms in all of cancer, and defects in phosphatase and tensin homologue (PTEN) tumor-suppressor activity are very often the cause (Vanhaesebroeck et al., 2012). The PTEN gene is deleted, mutated, and suppressed in cancers of various origins (Hollander et al., 2011). Pten alterations have been extensively studied in animal models of cancer, which collectively confirm that it is haploinsufficient (Berger et al., 2011a) for protecting from tumor initiation or progression along the kinase pathway (Kwabi-Addo et al., 2001; Trotman et al., 2003). Notably, PTEN protein has been found lost or low in many cancers in which its gene deletion is less frequent (Leslie and Foti, 2011). In full agreement, our comprehensive analysis at the DNA, RNA, and protein level revealed that more than half of the men who undergo radical prostatectomy present low or absent PTEN protein in spite of normal gene and gene expression status (Chen et al., 2011). Given the preeminent role of PTEN function in cancer, these findings suggest that interfering with PTEN protein degradation could greatly help many patients with cancer to stabilize disease, support therapy, and combat cancer progression. Defining gene loci that indirectly control major known tumor suppressors is a critical task. The function of the two major tumor suppressors besides PTEN, the p53 and retinoblastoma (RB1) proteins, has been shown to be tightly linked to the p16Ink4a and ARF proteins. Thus, the CDKN2 locus that encodes these proteins serves as a very important independent measure of p53 and RB1 function in cancer. Yet, no such indirect, PTEN-controlling tumor suppressors have so far been discovered.
Previous work has linked the polyubiquitination system to PTEN proteasomal degradation via the NEDD4-1 E3 ubiquitin (Ub)-ligase (Wang et al., 2007), and this role has been confirmed in vivo (Drinjakovic et al., 2010; Christie et al., 2012; Naguib and Trotman, 2013; Gupta et al., 2016). Yet, analysis of genes involved in PTEN ubiquitination has so far shown their rather infrequent alteration in cancer, suggesting that they are unlikely drivers of PTEN degradation in tumors with low PTEN protein. However, it has also emerged that discrete ubiquitination via this pathway can mediate PTEN nuclear import in cultured cells, as observed in animal models (Howitt et al., 2012; Naguib and Trotman, 2013; Goh et al., 2014) and a PTEN hamartoma syndrome family with inherited mutation of a PTEN ubiquitination site (Trotman et al., 2007). Active transport between the nucleus and cytoplasm is mediated by the importin/karyopherin system (Görlich and Kutay, 1999; Chook and Blobel, 2001). Import receptors recognize their cargo in the cytoplasm. The resulting complex can pass through the nuclear pore complex to reach the nucleus, where cargo is released. Cargo displacement is triggered by receptors binding to the GTP-bound form of RAN (Rexach and Blobel, 1995), a small GTPase protein of the RAS superfamily (Bischoff and Ponstingl, 1991). The importin–RAN-GTP complex shuttles back to the cytoplasm, where RAN-GTPase activation generates RAN-GDP, which falls off of the importin, thus enabling a new cycle of nuclear import.
Because this process could protect PTEN from degradation, we hypothesized that understanding PTEN stability requires understanding of its nuclear import. Thus, we sought to define the mechanism of PTEN nuclear transport and determine its role in controlling the PTEN levels in cancer.
Results
Importin-11 (IPO11) mediates PTEN nuclear import
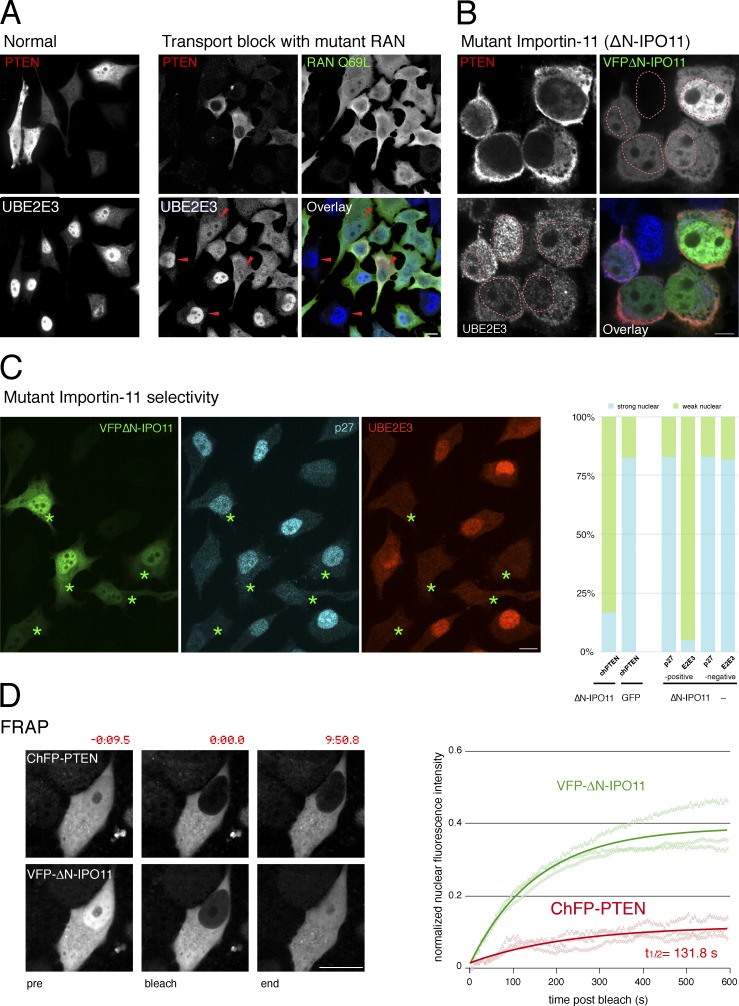
RAN-GTPase activity is essential for active nuclear transport (Melchior et al., 1993). Therefore, we first tested if catalytically dead RAN mutants interfere with PTEN import. As shown (Fig. 1 A and Fig. S1 A), PTEN was effectively excluded from nuclei of dominant-negative RANQ69L- or RANT24N-overexpressing cells, similar to a nuclear control protein, UBE2E3, and consistent with previous findings (Gil et al., 2006) confirming that PTEN uses an active import route. Unlike classical NLS-dependent transport, PTEN import depends on ubiquitination and is abolished by substitution of Ub-target lysines to arginine (Trotman et al., 2007). Because IPO11 is unique among import receptors in recognizing Ub-conjugated cargo (Plafker et al., 2004), we tested its role in PTEN transport. As shown (Fig. 1 B), the dominant-negative IPO11 mutant (ΔN-IPO11, a RAN-binding mutant, as shown in Fig. S1 B) caused strong cytoplasmic mislocalization of PTEN and the IPO11 cargo UBE2E3 (Plafker and Macara, 2000).
Figure 1.
Importin-11 mediates PTEN nuclear import. (A) Exogenous PTEN is both nuclear and cytoplasmic in PC3 (PTEN-null) cells (left, normal), and endogenous UBE2E3 is nuclear. Cells expressing the dominant-negative RANQ69L mutant reveal nuclear exclusion of exogenous PTEN and mislocalization of endogenous UBE2E3 (right, active transport block, arrowheads). Bar, 10 µm. Representative images of more than five independent experiments. (B) The dominant-negative ΔN-IPO11 mutant mislocalizes PTEN and a second IPO11 substrate, UBE2E3, in PC3 cells. Dashed red lines outline nuclei based on DAPI staining. Bar, 10 µm. Images represent >10 independent experiments. (C, left) Dominant-negative ΔN-IPO11 does not interfere with importin a/b–dependent nuclear import of p27Kip1 when it blocks import of the IPO11 target UBE2E3. Asterisks indicate ΔN-IPO11–positive cells. Bar, 10 µm. (right) Quantification of dominant-negative effects of ΔN-IPO11 on CherryFP-PTEN import (chPTEN + ΔN-IPO11, 54 cells, vs. chPTEN + GFP, 69 cells); P < 0.0001, two-tailed Fisher exact test and quantification of ΔN-IPO11 effect on p27 and UBE2E3 in 155 ΔN-IPO11–positive compared with 157 ΔN-IPO11–negative cells. See Fig. S1 C for all absolute and relative localization counts. (D) FRAP assays show that ΔN-IPO11 blocks PTEN nuclear transport, whereas the RAN-binding mutant importin still effectively shuttles. (left) Representative images of cells at pre- and postphotobleaching and at the experimental endpoints. Time stamps (m:ss.s) relative to bleach point are indicated. Bar, 10 µm. (right) Solid lines show recovery of mean nuclear fluorescence intensity over time in a typical experiment recording three individual cells (opaque lines) in the field of view.
The effect of dominant-negative IPO11 was selective as classic NLS: importin α/β–dependent nuclear import of p27 was unperturbed in cells in which the IPO11 cargo UBE2E3 was effectively mislocalized (Fig. 1 C, compare p27 and UBE2E3 in cells with asterisks; and see Fig. S1 C for quantification). We next used FRAP analysis in the few ΔN-IPO11–positive cells with nuclear PTEN to study transport dynamics as previously described (Trotman et al., 2007; Howitt et al., 2012). Photobleaching of nuclear mCherryFP-PTEN (ChFP-PTEN) in Venus-FP–ΔN-IPO11–positive (VFP-ΔN–IPO11) cells revealed no significant ChFP-PTEN recovery over ∼10 min, confirming that PTEN transport between the compartments is severely restricted (Fig. 1 D and Fig. S2, A and B). Note that a 10-min time frame has previously been demonstrated to suffice for import of UBE2E3 by IPO11 (Plafker et al., 2004) and for PTEN nuclear recovery in FRAP (Fig. S2 C; Trotman et al., 2007; Howitt et al., 2012). Furthermore, PTEN transport was independent of expression levels or cell size and functional in the presence of wild-type (wt) IPO11 (Fig. S2, A and C). The IPO11 mutant rapidly repopulated the nucleus with fluorescent VFP-ΔN–IPO11 from the cytoplasm, as expected for importins, which constantly shuttle between compartments. As shown by coimmunoprecipitation (coIP) assays, GFP-PTEN efficiently coimmunoprecipitated IPO11 (Fig. S2 D), and vice versa, exogenous IPO11 coprecipitated exogenous and endogenous PTEN (Fig. S2 E). Collectively, these data revealed that Importin-11 mediates and is required for PTEN nuclear import.
IPO11 regulates PTEN protein degradation
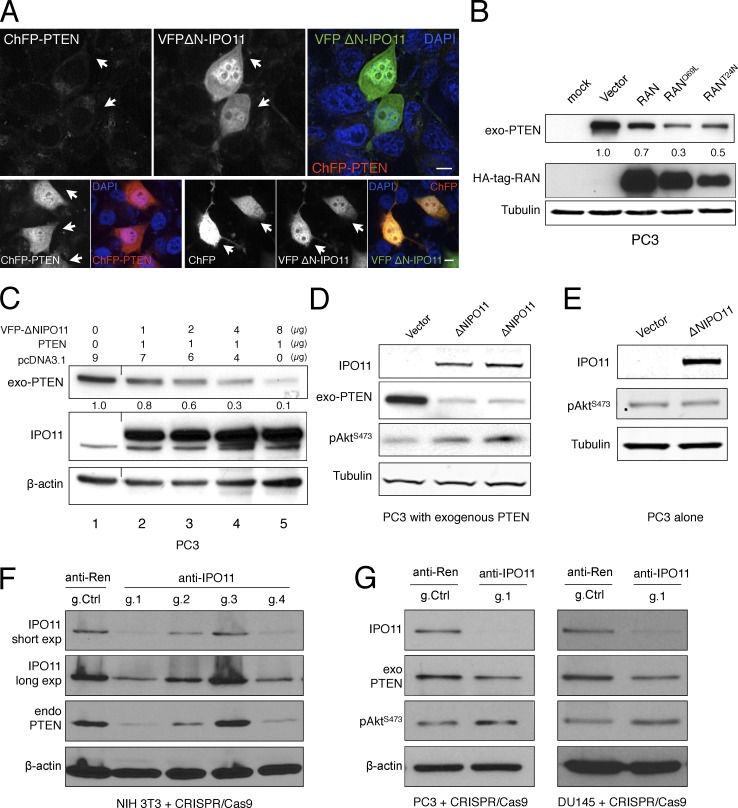
We noted that dominant-negative IPO11 significantly reduced the levels of ChFP-PTEN in a dose-dependent fashion, as did general interference with nucleocytoplasmic transport by RAN/mutant overexpression (Fig. 2, A–C). Whereas PTEN levels were lowered in a dose-dependent fashion by mutant IPO11 expression (Fig. 2 C), PTEN mRNA levels were not (Fig. S3, A and B). We consistently observed slight AKT activation when PTEN levels were lowered by the mutant importin (Fig. 2 D), but not in PTEN null cells (Fig. 2 E and Fig. S3 C). We confirmed these effects on both exogenous and endogenous PTEN by using RNAi and CRISPR-Cas9–based approaches to target IPO11 (Fig. 2, F and G; and Figs. S3 D and S7 D). Of note, we saw that the reduction in PTEN strongly correlated with strength of IPO11 suppression by the different CRISPR guides (Fig. 2 F). RNAi against Ipo11 lowered PTEN protein but not Pten mRNA (Fig. S3, E and F), and we next tested effects on PTEN ubiquitination. Cell treatment with the proteasome inhibitor MG132 indeed stabilized a pool of polyubiquitinated PTEN specifically upon expression of the IPO11 mutant, and CRISPR targeting of endogenous IPO11 increased PTEN ubiquitination when equal PTEN amounts were loaded (Fig. S4, A and B). Collectively, these data suggested that the dysfunctional import receptor triggers PTEN degradation in the cytoplasm.
Figure 2.
Importin-11 regulates PTEN protein degradation. (A) CherryFP-PTEN levels are diminished by coexpression with VFP-ΔN–IPO11 in PC3 cells (top, arrows) compared with expression alone (ChFP-PTEN alone, bottom left) or ChFP coexpression with VFPΔN-IPO11 (bottom right, arrows), and DAPI was shown in the overlay images (blue). Bars, 10 µm. Biological replicates (nbiol) = 4. (B) Blocking RAN-dependent nuclear import by overexpression of indicated HA-tagged mutants reduces PTEN levels (nbiol = 3). (C) Western blotting and quantification shows the dose dependence of ΔN-IPO11 effect on exogenous PTEN levels in PC3 cells (nbiol = 4). A black line indicates splicing of an unrelated column. (D) Transfection of the dominant-negative VFPΔN-IPO11 leads to AKT activation (p-AKT Ser473; nbiol = 3) in PTEN-transfected PC3 (PTEN-null) cells. Biological replicates are shown in lanes 2 and 3. (E) Dominant-negative VFPΔN-IPO11 has no effects on the p-AKT signaling pathway in the absence of PTEN in PC3 cells. nbiol = 2 (see Fig. S3 E). (F) CRISPR-Cas9–mediated knockout of Ipo11. Guide number 1 (g.1) shows the most efficient knockout of Ipo11, and this causes down-regulation of endogenous Pten compared with the control guide against firefly Renilla. Note that g.1 was designed to target both mouse and human IPO11. (G) Targeting of the IPO11 gene in PC3 (left) and Du145 (right) prostate cancer cell lines results in suppression of IPO11, which causes down-regulation of exogenous PTEN and activation of AKT.
IPO11 protects PTEN from the NEDD4-1/NDFIP1 lysine ubiquitination system
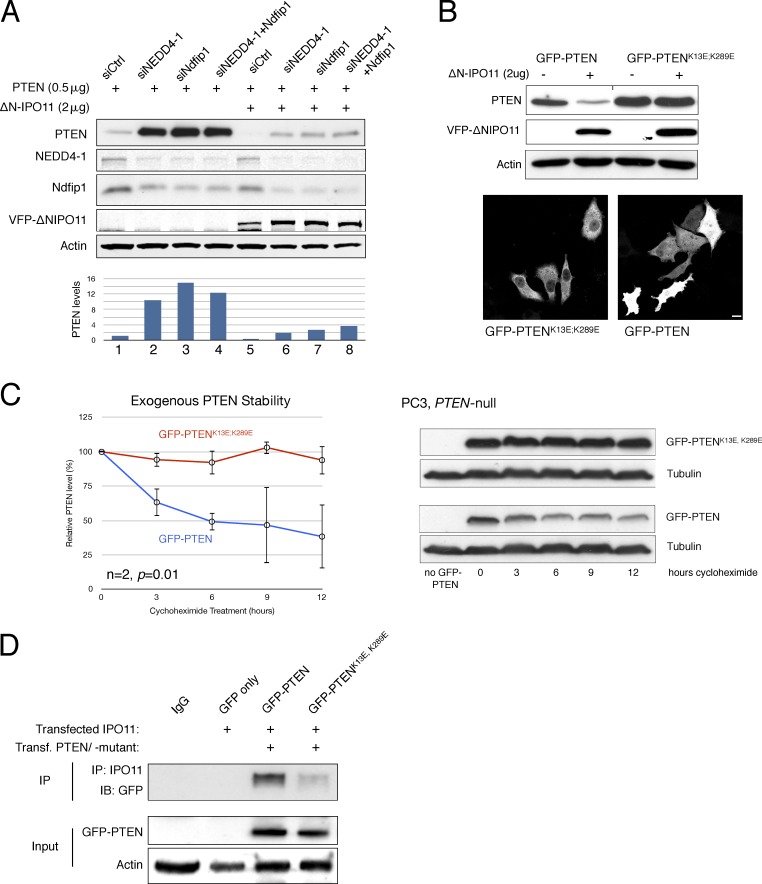
To expand on the aforementioned findings, we tested if known cytoplasmic agents of the PTEN ubiquitination system cooperate with mutant IPO11 (Trotman et al., 2007; Wang et al., 2007; Howitt et al., 2012). As shown (Fig. 3 A), knockdown of the NEDD4-1 E3-ligase, its adaptor protein NDFIP1, or both robustly increased PTEN levels. Most importantly, their knockdown completely restored PTEN protein to control levels in cells expressing mutant IPO11 (Fig. 3 A, compare PTEN expression in lanes 6–8 with lane 1). These data strongly suggested that the NEDD4/NDFIP1 system degrades PTEN upon IPO11 malfunction. Next, we found that the PTEN mutant harboring the combined human mutations of the known Ub-adduct site lysines (PTENK13E, K289E; Trotman et al., 2007) was largely insensitive to degradation induced by loss of IPO11 (Fig. 3 B, top). This was observed in spite of its strong cytoplasmic enrichment, shown in Fig. 3 B (bottom). Consistent with this result, the lysine mutant PTEN also showed an extended t1/2 relative to wt PTEN upon treatment of cells with cycloheximide (Fig. 3 C and Fig. S4 C). We further found that mutation of the PTEN ubiquitination sites interfered with IPO11 binding (Fig. 3 D), consistent with cytoplasmic mislocalization of this PTEN mutant (Fig. 3 B).
Figure 3.
Importin-11 protects PTEN from the NEDD4-1/NDFIP1 lysine ubiquitination system. (A) Western blot and quantification showing that VFP-ΔN–IPO11 regulates PTEN levels through the NEDD4-1– and NDFIP1-mediated ubiquitination pathway (biological replicates [nbiol] = 3). (B, top) Mutation of the PTEN Ub-adduct lysines (PTENK13E, K289E) rescues PTEN protein levels upon VFP-ΔN–IPO11 overexpression in PC3 cells. A black line in the PTEN row indicates splicing because of erroneous inverted loading. (bottom) The PTENK13E, K289E mutant is defective for nuclear import. Bar, 10 µm. nbiol = 4. (C) The t1/2 of GFP-PTENK13E, K289E (red line) is extended compared with that of GFP-PTEN (blue line) as shown by protein stability analysis using cycloheximide: quantification (left) of PTEN Western blots (right). P = 0.01. Error bars are SD of duplicates (see Fig. S2 F). (D) A coIP of IPO11 and GFP-PTEN/GFP-PTENK13E, K289E shows that mutation of PTEN Ub-adduct lysines impairs PTEN–IPO11 interaction in HEK293 cells (nbiol = 2). IB, immunoblot.
Collectively, our data revealed that disruption of IPO11 function leads to PTEN cytoplasmic degradation via NEDD4-1/NDFIP1 and two critical PTEN lysines that we previously identified as target sites for PTEN ubiquitination (Trotman et al., 2007).
UBE2E1 ubiquitinates PTEN and primes it for degradation
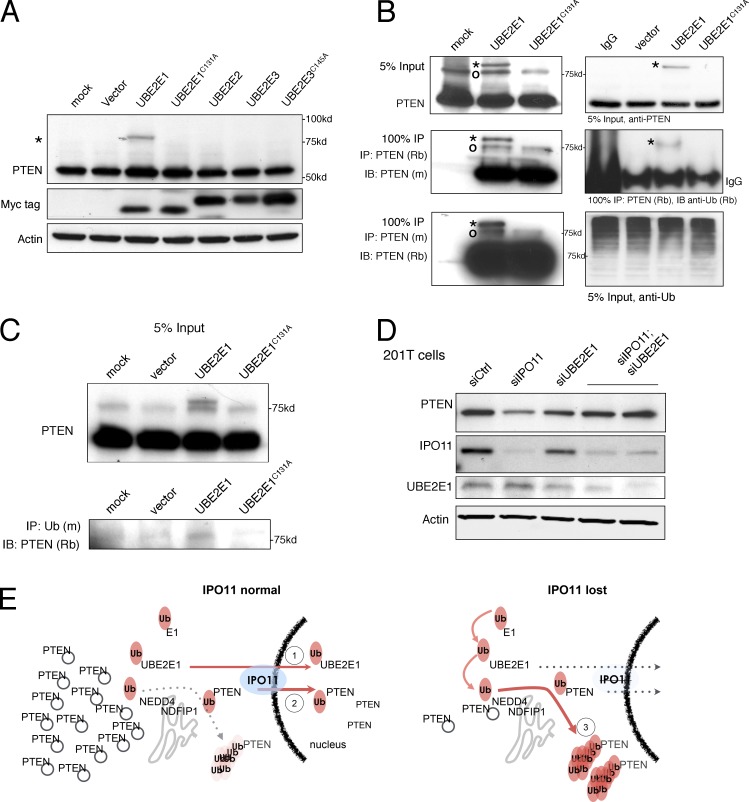
The results from Fig. 2 B suggested that general import blockade could destabilize PTEN far more effectively than cytoplasmic retention as obtained by fusion of PTEN to a nuclear export sequence (NES). This notion would be consistent with nuclear exclusion of a PTEN-degrading factor. NEDD4-1 contains a classic NES sequence (Hamilton et al., 2001), whereas NDFIP1 is a transmembrane Golgi protein (Harvey et al., 2002), thus firmly linking their combined action to the cytoplasm. Because our results furthermore specifically coupled IPO11 to PTEN degradation, we next tested IPO11-specific cargoes for their role in PTEN ubiquitination. Three Ub E2-conjugating enzymes (UBE2E1, -2, and -3) have been demonstrated to be imported by IPO11 when loaded with Ub (Plafker et al., 2004). UBE2E1 overexpression selectively modified endogenous PTEN with an adduct, whereas UBE2E2 and UBE2E3 did not (Fig. 4 A and Fig. S4 D, left). Furthermore, the active site cysteine mutant (Fig. 4 A, lane 4, C131A) demonstrated that active UBE2E1 is required for PTEN modification. Furthermore, in vitro reactions using recombinant proteins confirmed that incubation of PTEN with UBE2E1 produced PTEN adducts that were produced only by reactions including UBE2E1 (Fig. S4 D, right). To confirm that the adduct is PTEN derived, we performed PTEN IP/Western blotting with rabbit and mouse PTEN antibodies (Fig. 4 B, left). The discrete adduct band (Fig. 4 B, asterisk) appeared after UBE2E1 (but not mock or mutant UBE2E1C131A) overexpression. PTEN IP followed by staining with anti-Ub antibody confirmed this pattern (Fig. 4 B, right), and the reciprocal IP (Fig. 4 C, anti-Ub) followed by anti-PTEN Western blotting further confirmed that the adduct is Ub. Note that it remains unclear how many discrete Ub chains are added to PTEN. Most importantly, we found that UBE2E1 is also critical for the PTEN-degrading effect of IPO11. As shown in Fig. 2 (F and G), CRISPR-Cas9 or siRNA-mediated (Fig. S7 D) IPO11 targeting significantly lowered PTEN protein. This did not, however, alter PTEN mRNA levels (Fig. S3, E and F). In contrast, UBE2E1 knockdown had no major effect on PTEN protein (Fig. 4 D and Fig. S4 E, left). However, combined UBE2E1 and IPO11 knockdown rescued PTEN levels back to normal (Fig. 4 D). Furthermore, suppression of endogenous UBE2E1 also suppressed PTEN Ub adducts induced by MG132 treatment (Fig. S4 E, right). These data suggested that UBE2E1 is needed for PTEN degradation when IPO11 is impaired. Using a heterokaryon fusion assay, we confirmed that UBE2E1 rapidly shuttles between nucleus and cytoplasm, as seen previously for UBE2E3 (and unlike histone H2B; Fig. S4 F and Materials and methods). Note that we observed no bulk effect on PTEN localization upon UBE2E1 overexpression (not depicted), as only a small fraction of total PTEN was modified by UBE2E1 overexpression at steady state (Fig. 4 A).
Figure 4.
UBE2E1 discretely ubiquitinates PTEN and primes it for degradation. (A) Western blotting of endogenous PTEN reveals a slower migrating PTEN band (asterisk) >75 kD in HEK293 cells specifically upon UBE2E1 expression. Note that all UBE2E enzymes contain a Myc-tag (biological replicates [nbiol] = 4). (B) IP of PTEN from HEK293 cell lysates and immunoblotting (IB) with two different PTEN antibodies (6H2.1 mouse mAb [m] and a rabbit [Rb] mAb; see Materials and methods) confirms that adduct band (asterisks) is PTEN derived and cannot be formed by the catalytic dead UBE2E1C131A mutant. Note that a second PTEN-specific band is sometimes seen (circle); nbiol = 2. (C) Anti-Ub IP of lysates from HEK293 cells expressing wt or mutant UBE2E1 and anti-PTEN Western blotting (IB) confirms that the PTEN adduct is modified with Ub; nbiol = 3. (D) Knockdown of UBE2E1 rescues PTEN levels in the IPO11-depleted 201T cells (lanes 4 and 5; nbiol = 3). (E) Relay break model for the regulation of PTEN stability under normal (left, 1 and 2) and IPO11 mutant settings (right, 3). Note that NDFIP1 is cytoplasmic because it is a Golgi transmembrane protein and that NEDD4 contains a classical CRM1-type NES. Note that PTEN nuclear export is not depicted but is evident from, for example, FRAP analysis with cytoplasmic bleaching (not depicted). PTEN is illustrated on cytoplasmic vesicles (gray circles).
Based on our findings, we thus propose a relay break model that protects PTEN from degradation (Fig. 4 E). Functional Importin-11 breaks the cytoplasmic PTEN degradation machinery at two points (Fig. 4 E, left): (1) by importing, and thus sequestering, discretely ubiquitinated PTEN that has been primed for cytoplasmic polyubiquitination and degradation; and (2) by importing the activated Ub-loaded UBE2E1, thus limiting its ability to prime PTEN for degradation in the cytoplasm. Loss of IPO11, in contrast, fully exposes the Ub-primed PTEN to Ub-loaded, activated, and stranded cytoplasmic UBE2E1, thus completing the Ub relay system for PTEN degradation in concert with the cytoplasmic proteins NEDD4-1/NDFIP1 (Fig. 4 E, right). Notably, both nuclear import and PTEN degradation appear to require the same two Ub-adduct sites on PTEN, lysine 13 and lysine 289. Note that PTEN nuclear export is not depicted but always ongoing as is evident from, for example, FRAP analysis using cytoplasmic bleaching (unpublished data).
Ipo11 maintains Pten levels and nuclear localization to suppress cancer in mice
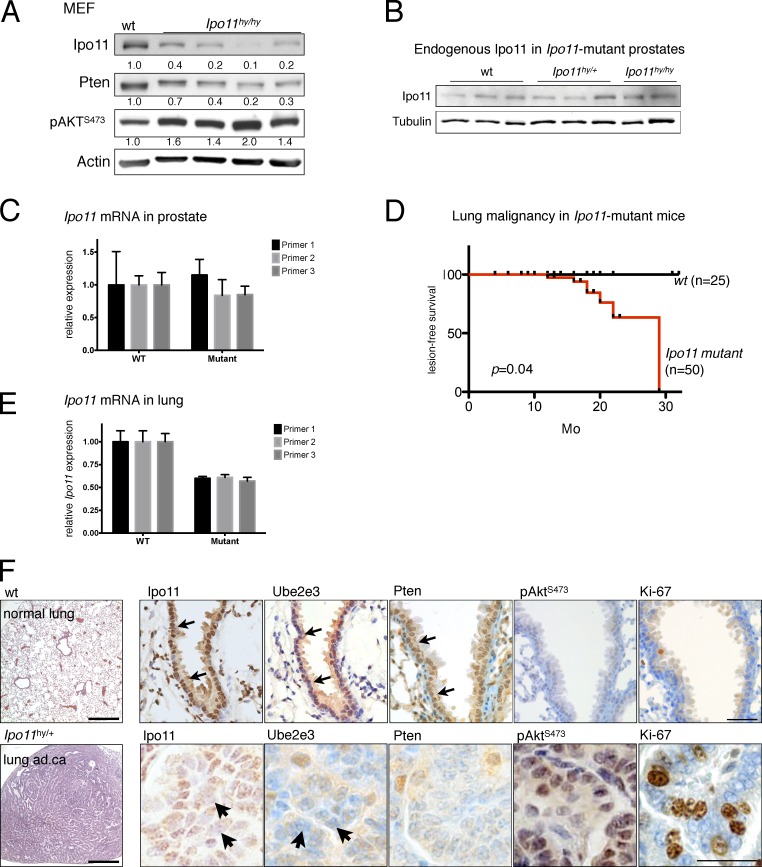
To determine if Importin-11 protects PTEN to prevent cancer, we generated Ipo11 mutant mice. These animals contain a strong splice acceptor site in the Ipo11 locus (Fig. S5, A–C; and Materials and methods). In primary homozygous mouse embryonic fibroblasts (MEFs), the resulting hypomorphic (hy) allele reduced Ipo11 mRNA to <25% of normal levels (Fig. S5 D). At the protein level, we found that, albeit some variability, Ipo11hy/hy MEFs had consistently lower Ipo11 levels (Fig. 5 A) compared with those of their wt counterparts. The changes in Ipo11 resulted in a correlating reduction of Pten protein and increased Akt activation (Fig. 5 A). Notably, Pten transcript levels were indistinguishable between normal and Ipo11 mutant cells, consistent with posttranslational deregulation of Pten (Fig. S5 D). These results from native, genetically engineered primary cells thus recapitulated the PTEN-protective role of IPO11 shown using cancer cell lines. Furthermore, we noted that Ipo11 mutant mice (hy/hy and hy/+ from hy/+ inter se breeding) are born at lower than expected Mendelian ratios, demonstrating partial embryonic lethality of the hy allele (n = 156; χ2 = 15.115; P = 0.0005; see Materials and methods). This finding strongly suggested a developmental threshold against survival of animals (and MEF cells) with very low Ipo11. Similar to these results, Pten hy mice and MEFs also show partial embryonic lethality and a strong bias against <25% of normal Pten protein levels (Trotman et al., 2003; Alimonti et al., 2010a). Of note, we found that CRISPR-based knockout of IPO11 in PTEN-deficient cells does not confer a growth advantage (rather a disadvantage), suggesting that, apart from PTEN, no major tumor-suppressive mechanism is suffering from loss of IPO11 (Fig. S5 D, right). However, we cannot exclude that Pten-independent Ipo11 functions are also essential in development.
Figure 5.
Ipo11 maintains Pten levels and nuclear localization to suppress lung cancer in mouse. (A) Western blotting of Ipo11 and Pten in wt and Ipo11hy/hy primary MEF cells (n = 4) confirms correlation of the endogenous Ipo11 and Pten proteins in vivo, as well as the subsequent activation of Akt (p-AKTS473). (B) Western blotting shows no Ipo11 protein reduction in the prostate lysates from 3-mo-old Ipo11 mutant mice of indicated genotypes. (C) Ipo11 mRNA levels are not suppressed by the splice trap in prostate as measured by quantitative PCR, suggesting that the trap is not functional in the tissue (biological replicates [nbiol] = 3). (D) Kaplan–Meier plot for lesion-free survival shows onset and penetrance of lung phenotypes in Ipo11hy/+ and Ipo11hy/hy mice (Fig. S5 E). Note that lymphomalignancy is not included. P-value (0.04) was calculated by log-rank (Mantel-Cox) testing. (E) Decreased transcription of Ipo11 is observed in preneoplastic 3-mo-old lungs from Ipo11hy/hy mice, suggesting successful knockdown (nbiol = 3). (F, top) H&E staining of wt lung and IHC analysis reveals (arrows) strong nuclear staining of Ipo11, nuclear Ube2e3 (confirming Ipo11 function), and Pten and weak cytoplasmic p-AktS473 staining. (bottom) Representative images of an Ipo11hy/+ lung adenocarcinoma reveals markedly reduced Ipo11 (arrows), mislocalized/reduced Ube2e3 (readout of impaired Ipo11 function; see arrows), and low/mislocalized Pten, but increased p-AktS473 and proliferation. See Fig. S6 C for more examples. Bars: (left) 500 µm; (right) 50 µm.
In adult Ipo11 mutant mice, we found that the Ipo11-splice trap did not work to suppress Ipo11 protein or mRNA in prostate (Fig. 5, B and C), consistent with lack of transcriptional interference in the Ipo11 locus in this tissue.
We then followed a cohort of 50 mutant and 25 wt mice with timed termination for analysis of neoplasia phenotypes and observed frequent adenocarcinoma, adenoma, hyperplasia, and neuroendocrine cancer of the lungs in these mice with onset at 12 mo (Fig. 5 D and Fig. S5 E). Using quantitative PCR, we confirmed that the splice trap efficiently suppressed Ipo11 mRNA in preneoplastic lung of 3-mo-old animals (Fig. 5 E). Histopathological examination revealed that adenocarcinoma showed predominantly papillary architecture with cytological atypia and increased proliferation (Figs. S5 F and S6 A). The high-grade neuroendocrine carcinoma consisted of small hyperchromatic cells with scant cytoplasm, negative for cytokeratin 5 (Fig. S6 A, neuroendocrine). Adenomas were well-circumscribed nodules measuring <5 mm and consisted of bland epithelial cells. Hyperplasia was defined as increase in number of cuboidal cells lining alveoli without cytological atypia, and atypical adenomatous hyperplasia lesions had atypical cuboidal cells lining the alveoli with mild atypia.
We could confirm causality and rule out off-target insertion effects of the splice trap through generation of a second Ipo11 mutant mouse cohort, which recapitulated the lung adenocarcinoma phenotype (see Materials and methods). Using immunohistochemistry (IHC) analysis on consecutive slides, we then tested the correlation between Ipo11 and Pten in lung (Fig. 5 F and Fig. S6, B and C). The lungs of wt animals showed predominantly nuclear Ipo11 staining. Ube2e3 staining (for which excellent IHC antibodies exist) showed strong nuclear localization, confirming functional Ipo11-mediated import in normal lung. Pten also showed strong nuclear staining in the epithelium (Fig. 5 F, top, arrows), whereas both phospho-Akt (p-Akt) and Ki67 were low. In contrast, Ipo11hy/hy lung adenocarcinoma showed only faint nuclear Ipo11 and Ube2e3 (Fig. 5 F, bottom, arrows), consistent with severely impaired Ipo11 function, as shown in Fig. 5 F and Fig. S6 (B and C) for different lesion types. Furthermore, Pten staining was low or absent, whereas Ki67 staining and p-Akt were increased. This pattern was consistently observed upon examination of adenoma, adenocarcinoma, and neuroendocrine lesions, as shown in Fig. S6 (B and C): loss of Ipo11 function, scored by Ube2e3 mislocalization, consistently correlated with substantially lowered Pten staining and some degree of (mostly nuclear) Akt activation.
Combined with our functional in vitro studies, these data suggested that dysfunctional Ipo11 lowers Pten levels by mislocalization in vivo. The long latency to tumor onset is consistent with the fact that the Ipo11 hy allele is not a complete null and with previous results on the long latency of Pten mutant lung cancer in mouse (Yanagi et al., 2007; Iwanaga et al., 2008).
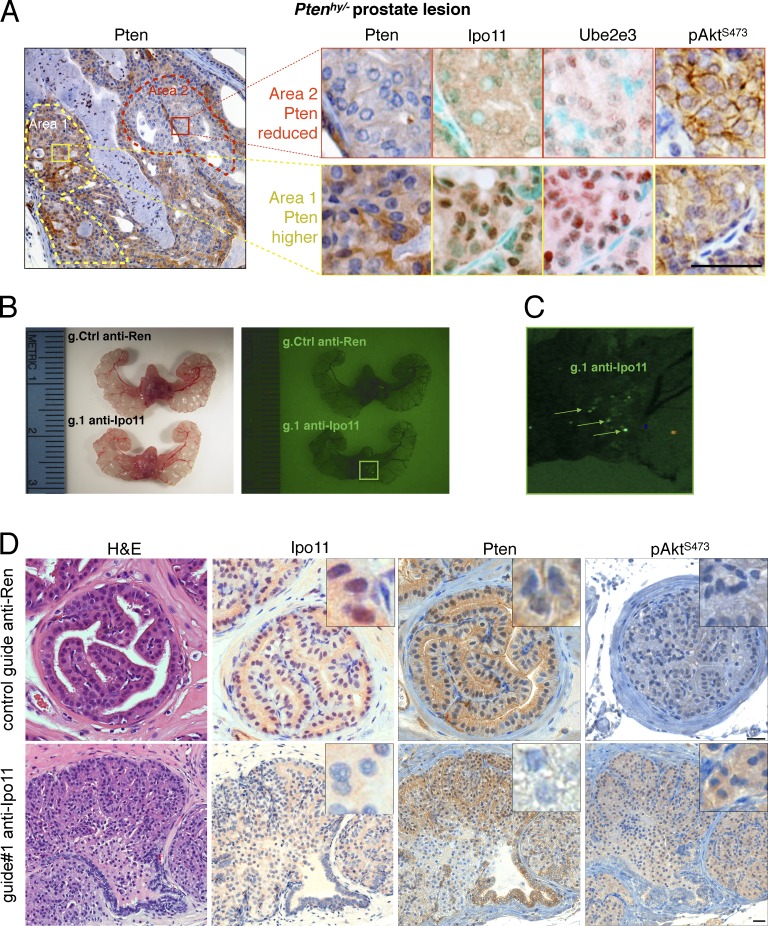
Ever since the discovery of PTEN, its deregulation has been observed in human prostate cancer (Li et al., 1997; Wang et al., 1998). These findings have been validated in mouse and today form the basis of genetically engineered mouse models for the most aggressive forms of prostate cancer (Podsypanina et al., 1999; Trotman et al., 2003; Chen et al., 2005; Cho et al., 2014). We had previously discovered that disease progression in Pten mutant prostate cancer is driven by the spontaneous, further suppression of Pten protein (Wang et al., 2007; Chen et al., 2011). However, the Ipo11 splice trap did not function to suppress Ipo11 in prostate tissue (Fig. 5 C). Thus, we first tested Ptenhy/− prostates, in which spontaneous Pten suppression is tightly correlated with Pten degradation and increased p-Akt and Nedd4-1 staining (Trotman et al., 2007). As shown in Fig. 6 A, the loss of Pten protein and classic p-Akt activation in prostate was tightly associated with loss of nuclear Ipo11 and Ube2e3 staining. Next, we examined Pten+/−; Phlpp1−/− prostates in which disease progression and Akt activation was also associated with spontaneous Pten suppression (Chen et al., 2011). As shown (Fig. S7 A), we found that dysfunction of Ipo11, as seen by cytoplasmic Ube2e3 mislocalization, correlated with marked reduction in Pten protein and activation of p-Akt. Collectively, our results confirmed the correlation between Ipo11 dysfunction and loss of Pten protein in prostate. This analysis also revealed that prostate tissue, with predominantly cytoplasmic and not nuclear Pten, suffers from decrease in Pten protein levels when Ipo11 is low.
Figure 6.
Importin-11 loss causes prostate neoplasia. (A, left) Typical Pten IHC of prostate from a Ptenhy/− mouse displays spontaneous reduction of Pten protein in area 2 (red dashes) compared with area 1 (yellow dashes). (right) Comparative IHC of Pten, Ipo11, Ube2e3, and p-AktS473 staining in area 2 (top) and area 1 (bottom) confirms correlation between Pten suppression and Ipo11 loss/malfunction as scored by Ube2e3 mislocalization. Note that methyl green (instead of hematoxylin) was used as nuclear counterstain in Ipo11 and Ube2e3 staining to reveal the faint remaining nuclear staining of Ipo11 and Ube2e3. Bar, 50 µm. (B) Comparison of mouse prostates 8 wk postinjection with lentiviral plasmids carrying Cas9, Venus (for fluorescence); control guide: anti-Ren (g.208); or guide targeting Ipo11: anti-Ipo11 (g.1) under light (left) and fluorescence (right). Fluorescence signals are not seen in anti-Ren–injected (g.208) animals. (C) Zoomed fluorescence picture of anti-Ipo11 injections show green signals (arrows) pointing to the sites at which virus injections led to the expression of Venus and genome-editing events. (D) IHC of prostate from lentivirus Cas9-CRISPR–injected mice shows neoplastic lesions with loss of epithelial architecture that was absent from Renilla control-targeted prostates. IHC analysis confirmed efficient Ipo11 suppression in anti-Ipo11–injected (bottom) but not anti-Ren–injected prostate (top). Comparative IHC of Pten, staining in anti-Ren control (top) and anti-Ipo11 tissue (bottom) confirms correlation between Pten suppression in regions with loss of nuclear Ipo11 and increased p-Akt tissue levels. Bars, 50 µm.
Finally, we tested if Ipo11 loss on its own causes suppression of Pten in prostate. To this end, we combined our viral injection–based RapidCaP Genetically Engineered Mouse system for prostate cancer (Cho et al., 2014, 2015; Nowak et al., 2015), with CRISPR-Cas9 knockout of Ipo11. 8 wk after intraprostatic injection of the CRISPR-Cas9 plasmids (anti-Ipo11 g.1), mice were sacrificed. The VFP transgene marker revealed sites of persistent expression of the Ipo11-targeting virus, which were not seen in the Renilla control-targeted prostates (Fig. 6, B and C; and Fig. S3 D for vector design). Hematoxylin and eosin (H&E) analysis (Fig. 6 D) of prostates from anti-Ipo11–injected mice revealed florid proliferation of neoplastic epithelial cells. The papillary tumors fill glands and are composed of uniform cells with round nuclei and moderate amphophilic cytoplasm. Cell borders are indistinct. At the margin of the papillary fronds, there are areas of multilayered cells with large basophilic nuclei. In contrast, control guide–injected mice presented normal prostate histology.
Next, IHC analysis confirmed efficient Ipo11 suppression, as revealed by the complete loss of nuclear Ipo11 protein expression. The adjacent sections presented with partial or complete loss of Pten and cytoplasmic activation of Akt. These results suggested that Ipo11 loss can cause neoplasia through suppression of Pten. In sum, we also noted that, regardless of Pten steady-state localization (cytoplasmic in prostate or nuclear in lung), loss or suppression of Ipo11 triggered an overall decrease in Pten levels, in agreement with our model (Fig. 4 E).
IPO11 protein correlates with PTEN protein levels in human lung cancer
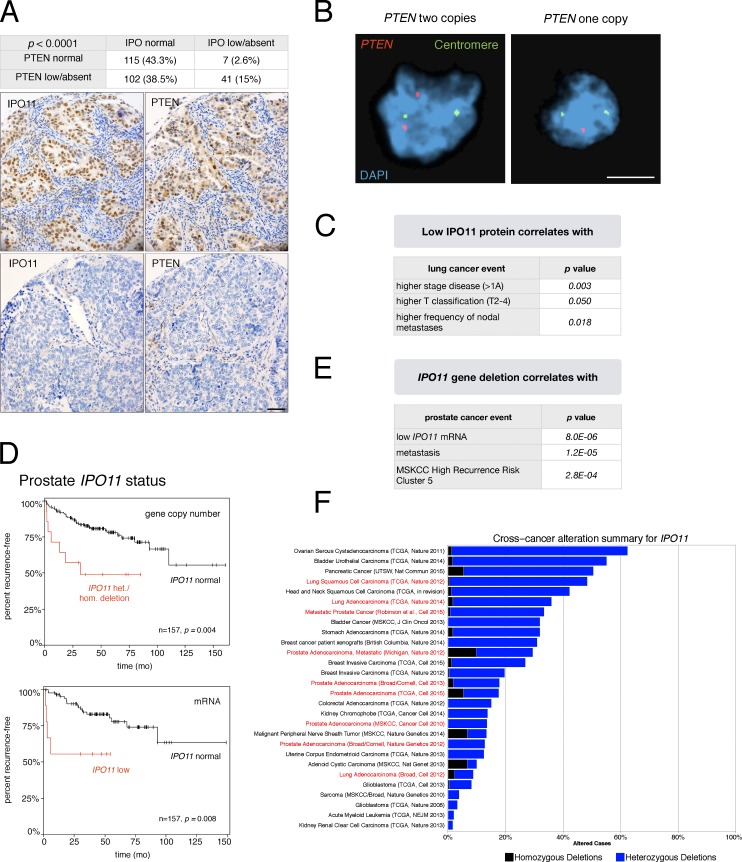
Based on the aforementioned results, we tested if IPO11 protein levels correlate with PTEN levels in human cancer. First, we studied tumor tissue microarrays (TMAs) of 265 lung cancer specimens (see Materials and methods; Wagner et al., 2011). This analysis revealed that PTEN protein is lower than normal in 54% of samples, similar to previous studies (Marsit et al., 2005; Scrima et al., 2012; Yanagawa et al., 2012). We found a highly significant correlation (P < 0.0001) between low or absent IPO11 and low or absent PTEN protein levels (Fig. 7 A). FISH analysis of PTEN DNA status on the TMAs confirmed that the majority of samples (94%) had two copies of PTEN (6% had hemizygous and none had homozygous loss, as illustrated in Fig. 7 B; Yanagawa et al., 2012). As outlined in the Discussion, these data suggested that loss of IPO11 may account for low PTEN presentation in approximately one-third of patients with lung cancer with low PTEN protein (60,000 patients in the U.S.; see Discussion). Furthermore, human IHC analysis confirmed our mouse results of overall decrease in PTEN levels in epithelial regions devoid of IPO11 (Fig. 7 A), consistent with the notion that IPO11 protects PTEN regardless of steady-state localization, along the model shown in Fig. 4 E. We next examined several lung cancer cell lines for IPO11 levels (Fig. S7 B) and found that PTEN protein levels correlated with those of IPO11 when contrasting the high- and low-level IPO11 cell lines (Fig. S7, B and C). Testing for the causal role of IPO11 in dictating PTEN levels in these cells, we found that both IPO11 knockdown and the dominant-negative VFP-ΔN–IPO11 strongly suppressed endogenous PTEN protein in the NCI-H2023 lung cancer cell line, consistent with our results from the 201T lung cancer cell line (Fig. S7 D). Collectively, our results suggested that IPO11 is critical for maintaining PTEN protein in human lung cancer. Analysis of the TMA-associated clinical data (Fig. 7 C) revealed that IPO11 absent/low-expressing tumors are associated with higher stage (higher than IA; 63 vs. 48%; P = 0.003), increased frequency of nodal metastases (27 vs. 13%; P = 0.018), and a trend to shorter overall survival, albeit not statistically significant (Fig. S7 E; P = 0.082). Multivariate analysis showed that low/absent IPO11 levels, when analyzed against other clinical factors, independently predicted advanced stage (stages II–IV; hazard ratio 2.3; confidence interval 1.14–4.63; P = 0.020), a strong predictor of survival.
Figure 7.
Importin-11 regulates PTEN protein levels in human cancer. (A, top) Statistical analysis of IPO11 and PTEN staining levels on the tumor TMA. (bottom) Representative example of correlation between low/absent IPO11 and PTEN levels. Bar, 50 µm. (B) Representative images of dual-color FISH of the samples from the TMAs using PTEN probe (red) and the centromeric probe of chromosome 10 (green), with DAPI counterstaining of the cell nuclei. (left) PTEN diploidy. (right) PTEN hemizygous loss. Bar, 50 µm. (C) Clinical correlates of low IPO11 protein staining derived from the cohort of 265 patients analyzed in A. (D) Kaplan–Meier outcome analysis for biochemical relapse after radical prostatectomy based on copy number (top) and gene expression changes (bottom) of IPO11 in the Memorial Sloan Kettering Cancer Center (MSKCC) patient with prostate cancer cohort (Taylor et al., 2010). (E) The table shows that IPO11 deletion is associated with high recurrence risk as previously defined for this dataset (Taylor et al., 2010). (F) IPO11 gene-deletion frequency across many cancer types. Homozygous and heterozygous deletions are shown. Note that lung, adeno, and squamous cell carcinoma are among the tissues with the most frequent IPO11 alteration.
In human prostate cancer, we found highly significant association between deletion of IPO11 with high risk of relapse after prostatectomy (biochemical recurrence), the most significant clinical parameter that separates the many men who live long after surgery from those who are likely to die of metastasis (Fig. 7 D, top). Importantly, low IPO11 mRNA expression was also associated with significant decrease in time to relapse, pointing specifically to the relevance of IPO11 among other genes deleted on chromosome 5q12 (Fig. 7 D, bottom). Furthermore, detection of IPO11 deletion in a surgical sample could identify men with high risk of metastasis (Fig. 7 F) and association with the previously published unbiased definition of the group of men with highest risk of relapse. Grouping was based on unsupervised clustering analysis of structural variation of the prostatectomy genomes (Taylor et al., 2010). Finally, our analysis of IPO11 status across all copy number alteration datasets curated at the cBioPortal (Fig. 7 F and Materials and methods) confirmed that patients with prostate and lung cancer are among the most likely to suffer from IPO11 gene deletion.
Discussion
PTEN levels are under tight control to balance growth restriction and survival. The Ub system has emerged as the prime regulator of PTEN levels, which need to be rapidly lowered upon insults that threaten cell survival, like ischemia or wounding (Naguib and Trotman, 2013). In spite of a wealth of data that link loss of PTEN protein to tumorigenesis, it has remained unclear which genetic alterations could indirectly and efficiently suppress PTEN (Leslie and Foti, 2011). Our results introduce the IPO11 nuclear import receptor as the Achilles’ heel of the checks and balances in the PTEN ubiquitination system (Fig. 4 E). Tissues, which rely on strong nuclear PTEN import, like lung, could be especially vulnerable to IPO11 failure. On top of maintaining PTEN levels, IPO11 could be equally important in supporting nuclear-specific PTEN functions that were previously reported (Song et al., 2012) or remain to be discovered. It is not clear at present how ubiquitinated PTEN interacts with Importin-11. The K13 and K289 sites, albeit residing in two different PTEN domains, are part of unstructured loops that could provide flexible, independently ubiquitinated interaction sites for the IPO11 transport receptor. This would be similar to the efficient interaction of importin-α with classical NLS signals that often work equally well regardless of N- or C-terminal fusion to a transgene and independently of transgene sequence.
Other relevant cancer genes may be co-lost with IPO11 in the broader 5q deletions, including the PI3K p85 subunit (PIK3R1 gene). PIK3R1 can increase kinase signaling both when lost and also when mutated as a gain of function (Taniguchi et al., 2010). Given our Ipo11-specific mouse results, it is tempting to speculate that the commonly observed codeletions of IPO11 and PIK3R1 could cooperate in many cancers. To date, 136 IPO11 missense mutations have been identified, 35 of them in lung cancer. Interestingly, only 19 PIK3R1 mutations have been identified in lung cancer among the 301 missense mutations found for the gene in all cancers (data from cBioPortal; see Materials and methods). IPO11 staining in human itself did not predict death or recurrence, yet our study did not have the power to do so. The fact that it did independently predict stage, which itself is the most powerful predictor of recurrence or death, is, however, indicative of the importance of IPO11 biology.
The frequent hemizygous IPO11 gene deletions (Fig. 7 F) are expected to suppress PTEN protein in PTEN diploid, as well as in PTEN hemizygous cells (Fig. 2 G), and thus could be very effectively contributing to tumor initiation or progression, respectively. Loss of Pten has been shown to contribute to Ras-driven lung cancer in mice (Iwanaga et al., 2008). However, PTEN deletion is typically hemizygous in patients with lung cancer and has been reported to be some 10 times less frequent than loss of the protein (Marsit et al., 2005; Scrima et al., 2012; Yanagawa et al., 2012). IPO11 maintains PTEN protein in a dose-dependent fashion, hence IPO11 heterozygous loss may already contribute to cancer via the PI3K pathway, a well-known driver of lung cancer (Castellano and Downward, 2010; Wojtalla and Arcaro, 2011). We have found that 15% of patients presented with low IPO11 and PTEN protein but two copies of the PTEN gene (Fig. 7, A and B). Given a population of 409,000 patients with lung cancer in the U.S. alone (Howlader et al., 2016), >60,000 of them would fall into the category in which PTEN function might be restored by interfering with degradation.
In prostate, the role of the PTEN/PI3K pathway is very well established. More than a decade’s worth of animal model–based identification of the key pathway lesions (Irshad and Abate-Shen, 2013) has now seen effective confirmation in the human prostate cancer genome studies (Liu et al., 2009; Taylor et al., 2010; Grasso et al., 2012; Cancer Genome Atlas Research Network, 2015; Robinson et al., 2015). Although the overall degree of structural DNA variation has emerged as the strongest predictor of disease progression (Hieronymus et al., 2014), these studies have so far not pointed to any distinct genetic events that can predict progression to lethal prostate cancer. Importantly, most primary cancers that require intervention reveal loss of PTEN protein (Carver et al., 2009; Chen et al., 2011). Yet the PTEN gene is most frequently deleted only later, in a mean 54% lethal therapy-resistant metastatic samples (Taylor et al., 2010; Grasso et al., 2012; Robinson et al., 2015). It has been shown that complete PTEN loss and strong AKT activation can trigger senescence and immune responses that delay or prevent prostate cancer progression when p53 is intact (Chen et al., 2005, 2011; Alimonti et al., 2010b; Nardella et al., 2011; Di Mitri et al., 2014). We therefore infer that the degradation of PTEN protein observed after loss and impairment of Importin-11 is a very effective driver of human prostate cancer progression.
We conclude that Importin-11 function is integral to PTEN stability. Thus, our work introduces a novel locus that could be an important target of the chromosome 5q deletions seen in many solid tumors beyond prostate and lung cancer.
Materials and methods
Mice
Two Ipo11 embryonic stem (ES) cell lines (129/Ola), RRR432 and RRU483, were obtained from Bay Genomics. RT-PCR using the reverse primer (P2) 5′-GACAGTATCGGCCTCAGGAAGATCG-3′, RRU483 forward primer (P1) 5′-TTCTACCATGTCACCAAGAC-3′, and RRR432 forward primer 5′-TCAGGATACTGCTGTGTTGA-3′ was applied to confirm the insertion of the gene-trap vector βGEO in introns 1 and 5, respectively. The resulting single band was further confirmed by restriction digest with PvuII, which gave rise to a 147-bp fragment off the βGEO cassette. Then two independent Ipo11 hy lines were generated by microinjection into host blastocysts using standard procedures. The chimeric mice were bred to C57BL6 mice, and resulting Ipo11 mutant mice (129/C57BL6) were crossed with C57BL6 wt for three generations. Those offspring were intercrossed to obtain the study cohort. The mouse line derived from the RRU483 ES cells was fully examined and shown throughout this manuscript. For genotyping of the RRU483 mouse line, PCR primers (P3–P5) 5′-CGCGCGGGCACTGTATAAGTC-3′, 5′-AGCTACTTGGGAACTGTGCTGGGC-3′, and 5′-ATGAGGGAGCAGGGCTCGGC-3′ were used. To test for Mendelian genotype ratios, we followed 23 litters resulting from heterozygous (Ipo11hy/+) intercrosses. The expected ratio of wt to heterozygous to homozygous animals is 1:2:1, and we obtained 60 wt (29 males and 31 females), 65 heterozygous (33 males and 32 females), and 31 homozygous (15 males and 16 females) animals, demonstrating a significant selection against carriers of the hy allele. 50 animals were followed up, and groups of two to three animals were sacrificed in 2-mo intervals. Disease-free survival (DFS) curves were calculated using the Kaplan–Meier method and log-rank (Mantel-Cox) testing with Prism 5.0 (GraphPad Software) for Macintosh OS X (Apple).
CRISPR-Cas9 plasmids
The guide RNAs (gRNAs) were designed as previously described, and the highest-ranking gRNAs were chosen (Nowak et al., 2015). Anti-IPO11 guides were: g.1, 5′-CACAGCAGTATCCTGACTGG-3′; g.2, 5′-CCCTTGATATAAATGTACGG-3′; g.3, 5′-ATGGGATTGATCGTTACTGG-3′; and g.4, 5′-GAAAAGTCAACATTGCGTGC-3′. The gRNA oligonucleotides were purchased from Sigma-Aldrich, phosphorylated, and annealed according to previously published protocols (Cong et al., 2013). The duplex oligonucleotides were subsequently cloned into the lentiCRISPRv2 plasmid (plasmid 52961; Addgene), replacing the 2-kb filler as previously described (Sanjana et al., 2014; Nowak et al., 2015).
Cloning CRISPR-Cas9 venus plasmids (ECPV)
lentiCRISPRv2 plasmid was acquired from Addgene (plasmid 52961; Sanjana et al., 2014). lentiCRISPRv2 plasmid was modified to use Venus fluorescent protein. BamHI cut after Cas9 plasmid not containing P2A and puromicine cassette was created. First, we excised the P2A-Puro–WPRE-3′LTR part with BamHI/PmeI restriction enzymes, religated a PCR product containing only the WPRE-3′LTR piece, surrounded by BamHI/PmeI, and ligated it to already cut with BamHI/PmeI lentiCRISPRv2. We amplified PCR product P2A-Venus surrounded by BamHI sites from both sites and cloned to the BamHI-containing plasmid in the correct orientation; our vector is named ECPV, and lentiCRISPRv2, for nomenclature and consistency purposes, is called ECPP.
Lentivirus plasmid production and infections
For in vivo injections, lentiviruses were produced and concentrated as previously described. Anti-Ipo11 virus–infected animals were generated by intraprostatic delivery of ECPV viruses and analyzed as previously described (Nowak et al., 2015).
For in vitro experiments, cells were washed with 1× PBS and infected with freshly collected lentivirus (ECPP backbone) at day 0. 24 h postinfection, the medium was changed for medium containing puromycin at 3 µg/µl (day 1). At day 4, cells were washed with 1× PBS, and fresh antibiotic was added for another 3 d. At day 7, the medium was replaced with 10% FBS/1× PBS DMEM (without antibiotic) to allow cells to recover. Cells were analyzed at day 12 after infection by Western blotting to assess IPO11 protein knockout. As the initial screen for the knockout of IPO11 at the protein level was very efficient without requiring single-cell clonal expansion, we focused on using and analyzing the polyclonal population.
Transfection plasmids
For transfection experiments, the plasmids pmCherry-C1-RanQ69L (plasmid 30309; Addgene; Kazgan et al., 2010) and pTK21 (plasmid 37396; Addgene; Kiyomitsu and Cheeseman, 2012) were purchased.
Patients and clinicopathological data
The TMA was constructed from a cohort of 402 patients with non–small-cell lung cancer operated on at New York–Presbyterian Hospital, Weill Cornell Medical Center, between 1992 and 2007, including 302 consecutive and uniformly treated tumors resected between 2005 and 2007. The study was approved by the New York–Presbyterian Hospital/Weill Cornell Medical Center Institutional Review Board. Patients with the following characteristics were excluded from analysis: multiple tumors, stage IV disease, recurrences from previous tumors, preoperative nonsurgical therapy, and incomplete staining. Patient demographics, clinical and pathological staging, and follow-up information were obtained from a prospectively maintained, institutional thoracic surgery registry.
The final TMA included 265 patients: median age 69 y, 60% females, and 80% current or former smokers. Among the patients, the majority of tumors (75%) were stage IA (50%) or IB (25%), whereas the remainder were stage II (8%), III (13%), or IV (4%). Most tumors (76%) were subtypes of adenocarcinoma. Cox proportional hazards regression analysis was done to determine the independent effects of clinical predictors on the likelihood of nodal metastases, on advanced stage (II–IV), and the effect on DFS. Variables included in the model were: age, gender, smoking status, tumor size, tumor differentiation, and histological subtype. Univariate predictors with probability values <0.20 were included in the multivariate model. Survival probabilities were estimated using the Kaplan-Meier method. The log-rank test was used to determine significance of survival distributions among groups. Data analysis was performed using IBM SPSS software (version 22.0; SPSS Inc.).
Tumor TMAs and FISH
For analysis of protein status, PTEN and IPO11 IHC was performed on a lung cancer tumor TMA with 265 evaluable cases in triplicate including normal tissue controls. PTEN and IPO11 staining was scored for each case as 0, +1, or +2 by two reviewers using consensus scoring. All patient sample analyses were performed on deidentified patient data and material and thus qualified for exemption from human subject statements under 45 CFR 46.101 (b). To assess the gene status of PTEN by FISH analysis, a locus-specific probe and reference probe were used, as previously described (Berger et al., 2011b).
Western blotting
Cells and lungs from dissected animals of all genotypes and cell lines were homogenized and lysed in 50 nM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% NP-40, 1 mM sodium orthovanadate (Na3VO4), 10 mM NaF, and protease inhibitor cocktail (Roche) and cleared by centrifugation; concentrations were determined by protein assay (Bio-Rad Laboratories), and samples were combined with SDS sample loading buffer followed by brief sonication and centrifugation. The supernatant was collected for Western blotting. Western blotting was done using the following polyclonal antibodies from Cell Signaling Technology: Myc-tag (#2272), p-Akt Ser473 (#4051), p-S6 ribosomal protein Ser235/236 (#4856), and p-PRAS40 Thr246 (#2997), as published. Other polyclonal antibodies used were IPO11 (AP9661b; Thermo Fisher Scientific), UBE2E1, UBE2E3, NEDD4-1 (07-049; EMD Millipore), Ndfip1 (HPA009682; Sigma-Aldrich), and Ub (P4D1; Santa Cruz Biotechnology, Inc.). mAbs used were HA-tag (12CA5; homemade), β-actin (A5441; Sigma-Aldrich), γ-tubulin (T6199; Sigma-Aldrich), GFP (JL-8; Takara Bio Inc.), and c-Myc (ab32072; Abcam). Western blots were developed using the enhanced chemiluminescence detection reagent (EMD Millipore) or the Odyssey Infrared Imaging System (LI-COR Biosciences). Quantification was done using the LI-COR system or densitometry using ImageJ 1.38X software (National Institutes of Health).
In vitro ubiquitination assay
In vitro ubiquitination assay was performed by mixing 0.25 µg E1, 8 µg Ub, 3 µg E2, 40 ng PTEN (R&D Systems), 5 µM Ub aldehyde, 150 mM NaCl, 50 nM ZnCl2, and Energy Regenerating System in a 20-µl volume with or without 10% rabbit reticulocyte lysate at 37°C for 4 h. Reaction was stopped by the addition of 2× Laemmli buffer and boiling. The resulting lysates were subjected to SDS-PAGE, transferred to nitrocellulose, and probed for PTEN.
IHC, immunofluorescence (IF), and quantifications
Tissues were fixed in 10% formalin and embedded in paraffin. Sections were processed with H&E reagents or stained for PTEN/MMAC1 Ab-2 (#RB-072-PO; NeoMarkers), Ki-67 (VP-K451; Vector Laboratories), Cytokeratin 5 (AF-138; Covance), IPO11 (AP9661b; Thermo Fisher Scientific), and UBE2E3 using the Discovery XT processor system (Ventana Medical Systems, Inc.) as previously described. The slides were processed using citrate buffer–based (10 mM, pH 6.0) antigen retrieval, and the avidin–biotin peroxidase IHC method. Ki-67–stained cells from five random fields of a lesion were counted after imaging with an Axioplan microscope (40× objective; ZEISS) to quantify cell proliferation. Statistical analysis was performed using the one-tailed Student’s t test. For IF, cells were seeded on glass coverslips in 24-well plates and fixed with 4% PFA for 10 min followed by 50 mM NH4Cl for 10 min, 0.5% Triton X-100 for 5 min, and PBS (washing three times), 10% goat serum (30 min), and primary antibodies HA (clone 12CA5; Cold Spring Harbor Laboratory Antibody Facility), UBE2E3, PTEN (6H2.1; Cascade BioScience), p-Akt (Ser473, IHC-specific; #4060; Cell Signaling Technology), NEDD4-1 (07-049; EMD Millipore), and Myc-tag (#2272; Cell Signaling Technology) were applied for 5 h at 4°C or as indicated by the manufacturer. Secondary antibodies were goat anti–rabbit and goat anti–mouse Alexa Fluor 546, Alexa Fluor 488, and Cy5 (Invitrogen/Molecular Probes), and DAPI (1:500; Sigma-Aldrich) was used to counterstain cell nuclei. Coverslips were mounted using ProLong Gold Antifade reagent (P36930; Invitrogen) and sealed with clear nail polish after curing at RT. Confocal analysis was performed on a spinning disk confocal microscope (PerkinElmer) using Volocity software v. 5.3.2. For quantification of PTEN localization, nuclear and cytoplasmic intensity ratios were calculated and averaged for 10 cotransfected cells of each condition.
Heterokaryon fusion assay
HeLa cells were transfected by the calcium phosphate method with plasmids encoding Myc-UBE2E1, Myc-UBE2E2, or H2B-RFP. 1 d posttransfection, cells were mixed a separate HeLa cell line stably expressing H2B-GFP. The cell mixtures were reseeded onto coverslips and grown for 24 h. The next day, cells were fused by incubation with 50% polyethylene glycol 8000 in Hanks’ buffered saline for 2 min at RT. After extensive washing, the cells were incubated at 37°C for 2 h in the presence of 35 µM cycloheximide to block the translation of new proteins. Cells were then fixed and permeabilized in preparation for IF analysis. The Myc tag was visualized with anti-Myc IF, RFP and GFP were visualized by epifluorescence, and nuclear DNA was counterstained with Hoechst. Photomicrographs were captured with a 60× objective.
Cell culture
MEFs were isolated as previously described from individual embryos of various genotypes and cultured in 10% FBS in DMEM (with antibiotics and glutamine). Cells were plated in triplicates at 1.25 × 104 cells/well in 12-well plates for determination of growth curves by spectroscopic measurement of crystal violet uptake. For t1/2 studies, cells were incubated in the presence of 25 µg/ml cycloheximide for the indicated times. siRNA SMARTpool (GE Healthcare) against IPO11, UBE2E1, UBE2E2, UBE2E3, NEDD4-1, and NDFIP1 was transfected by DharmaFECT3 (GE Healthcare) using siRNA SMARTpool against GAPDH as controls. Cells were harvested and subjected to Western blot, IF, and quantitative real-time PCR (qRT-PCR) 96 h after transfection. FACS procedures were performed on an FACSAria II system (BD). Cells were harvested 18 h posttransfection. The single-cell suspensions were prepared using a standard trypsinization protocol and resuspended in media that is calcium and magnesium free and contains no phenol red. 5 × 106 cells was filtered and sorted into round-bottom tubes containing cell media (#2054; BD). Representative sorting gates are shown in Fig. S7 D. For MG132 (EMD Millipore) treatment, at 18 h after transfection, cells were treated with 20 µM MG132 for 4 h. Cells were treated with 20 µM leptomycin B (#9676; Cell Signaling Technology) for 3 h at 18 h posttransfection.
qRT-PCR
RNA was isolated from tissue or cells using the TRIzol method, and cDNA was produced from 2 µg RNA using the SuperScript III system with oligo dT primers (Invitrogen) as suggested by the manufacturer. qRT-PCR was performed on the LightCycler 480 Real-Time PCR System using the SYBR Green I Master (Roche) and the following amplification protocol: 5 min at 95°C for 40 cycles (15 s at 94°C, 10 s at 60°C, and 10 s at 72°C) followed by determination/confirmation of amplicon melting temperature. Reactions were performed in triplicates; three primer pairs for each gene were confirmed to yield a single amplicon band by 2% agarose gel electrophoresis. Absence of amplification from non–reverse-transcribed RNA was confirmed to exclude genomic DNA amplification. Quantifications were done using LightCycler 480 Relative Quantification Software (Roche). The mouse primer sets used were obtained from PrimerBank (http://pga.mgh.harvard.edu/primerbank): Ipo11 (PrimerBank ID: 31541898a), forward (Fwd) 1, 5′-TGTGCCAGTCCTTTTATGAGC-3′ and reverse (Rev) 1, 5′-CCTCTCGCCTTCAATAACACCT-3′; Fwd2, 5′-TGAATCCAGTATTAGGCCCACA-3′ and Rev2, 5′-CCTCTCGCCTTCAATAACACCT-3′; Fwd3, 5′-CAGGTTTGTGCCAGTCCTTTT-3′ and Rev3, 5′-TGGATACCTCTCGCCTTCAATA-3′; Pten (PrimerBank ID: 6679523a), Fwd1, 5′-TGGATTCGACTTAGACTTGACCT-3′ and Rev1, 5′-GCGGTGTCATAATGTCTCTCAG-3′; Fwd2, 5′-TGCACAGTATCCTTTTGAAGACC-3′ and Rev2, 5′-GAATTGCTGCAACATGATTGTCA-3′; Fwd3, 5′-CAATCATGTTGCAGCAATTCACT-3′ and Rev3, 5′-ATCTAGGGCCTCTTGTGCCTT-3′; and Hprt1 (PrimerBank ID: 7305155a), Fwd, 5′-TCAGTCAACGGGGGACATAAA-3′ and Rev, 5′-GGGGCTGTACTGCTTAACCAG-3′. The human primer sets used were as follows: Actin (PrimerBank ID: 4501885a1): Fwd, 5′-CATGTACGTTGCTATCCAGGC-3′ and Rev, 5′-CTCCTTAATGTCACGCACGAT-3′; IPO11 (PrimerBank ID: 198041776c1), Fwd1, 5′-CAGGCCACCAGTCAGGATACT-3′ and Rev1, 5′-GAGAGAGCATGAGGTGCTACA-3′; Fwd2, 5′-AAGTTGCTAGATTGGATTGTCCC-3′ and Rev2, 5′-GCAAGTCGTTTAGATGCCAGT-3′; PTEN (PrimerBank ID: 110224474c1), Fwd1, 5′-TGGATTCGACTTAGACTTGACCT-3′ and Rev1, 5′-GGTGGGTTATGGTCTTCAAAAGG-3′; Fwd2, 5′-TTTGAAGACCATAACCCACCAC-3′ and Rev2, 5′-ATTACACCAGTTCGTCCCTTTC-3′; and Neomycin, Fwd1, 5′-GGCTATGACTGGGCACAACA-3′ and Rev1, 5′-CGCTGACAGCCGGAACAC-3′; and Fwd2, 5′-GCGGCGGCTGCATACGCTTGATCCGG-3′ and Rev2, 5′-CGCCATGGGTCACGACGAGATCC-3′.
Data collection and statistical considerations
Database and hospital records were reviewed for each patient for demographic, clinical, and pathological data. Staining was correlated to clinical and pathological factors using χ2 analysis. Median follow-up time was calculated on the basis of surviving patients. Overall survival and DFS were estimated with the Kaplan–Meier method from the date of surgery with significance determined by log-rank. Overall survival was estimated from the date of surgical resection until death from any cause or the date of last follow-up. DFS was estimated from the date of surgical resection until tumor recurrence or death from any cause. All p-values were two-sided. Cox proportional hazards regression analysis was used to identify univariate predictors of survival. All univariate predictors with a p-value ≤0.2 were entered into a multivariate Cox proportional hazards regression model of factors predicting survival. The statistical package used was SPSS statistical software version 17.0 (SPSS Inc.). Whole-genome copy number alteration frequencies were derived from the studies curated at the cBioPortal for Cancer Genomics (http://www.cbioportal.org/public-portal/) using the following query: DATATYPES: HOMDEL HETLOSS; IPO11 on all samples. IPO11 and PIK3R1 mutation analysis was done using the query: DATATYPES: MUT; IPO11 and DATATYPES: MUT; PIK3R1 on all samples and on the lung tumor subset. Analysis of deletion sizes in the IPO11 locus, outcome, and correlation analysis for prostate cancer was done using the Nexus Copy Number software version 8.0 (BioDiscovery) with the Memorial Sloan Kettering Cancer Center dataset (Taylor et al., 2010), as published previously (Chen et al., 2011).
Online supplemental material
Fig. S1 shows PTEN nuclear import is RAN dependent. Fig. S2 shows PTEN nuclear import is blocked by a dominant-negative mutant of IPO11. Fig. S3 shows validation of transgenic, knockdown, and knockout tools. Fig. S4 shows functional analysis of UBE2E1 perturbation. Fig. S5 shows genetic engineering and analysis of the Ipo11 gene trap mouse. Fig. S6 shows histopathology analysis of malignancy in the Ipo11 gene trap mouse. Fig. S7 shows correlation of IPO11 and PTEN in cells, mouse prostate, and human lung cancer.
Supplementary Material
Acknowledgments
The authors thank J. Hicks, and N. Schultz for discussion and help with genome analyses; L. Bianco, A. Nourjanova, K. Manova, and A. Barlas for help with histology procedures and analysis; P. Moody for help with FACS procedures; and D. Tsang for help with the manuscript.
L.C. Trotman is a Research Scholar of the American Cancer Society and is supported by the Pershing Square Sohn Foundation and the U.S. Department of Defense (W81XWH-13-PCRP-IDA). This work was also supported by the American Cancer Society (RSG-14-069-01-TBE to L.C. Trotman), the National Institutes of Health (CA137050), the Robertson Research Fund of Cold Spring Harbor Laboratory, and the National Institutes of Health to the Cold Spring Harbor Laboratory Cancer Center through support grant 5P30CA045508 for funding of microscopy, flow cytometry, gene sequencing, and animal husbandry. S.M. Plafker received support from the National Institute of General Medical Sciences (1R01GM092900-03).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ChFP
- mCherryFP
- coIP
- coimmunoprecipitation
- DFS
- disease-free survival
- ES
- embryonic stem
- gRNA
- guide RNA
- H&E
- hematoxylin and eosin
- hy
- hypomorphic
- IF
- immunofluorescence
- IHC
- immunohistochemistry
- MEF
- mouse embryonic fibroblast
- NES
- nuclear export sequence
- PI3K
- phosphoinositide 3-kinase
- PTEN
- phosphatase and tensin homologue
- qRT-PCR
- quantitative real-time PCR
- TMA
- tissue microarray
- Ub
- ubiquitin
- VFP
- Venus-FP
- wt
- wild type
References
- Alimonti A., Carracedo A., Clohessy J.G., Trotman L.C., Nardella C., Egia A., Salmena L., Sampieri K., Haveman W.J., Brogi E., et al. . 2010a Subtle variations in Pten dose determine cancer susceptibility. Nat. Genet. 42:454–458. 10.1038/ng.556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alimonti A., Nardella C., Chen Z., Clohessy J.G., Carracedo A., Trotman L.C., Cheng K., Varmeh S., Kozma S.C., Thomas G., et al. . 2010b A novel type of cellular senescence that can be enhanced in mouse models and human tumor xenografts to suppress prostate tumorigenesis. J. Clin. Invest. 120:681–693. 10.1172/JCI40535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A.H., Knudson A.G., and Pandolfi P.P.. 2011a A continuum model for tumour suppression. Nature. 476:163–169. 10.1038/nature10275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M.F., Lawrence M.S., Demichelis F., Drier Y., Cibulskis K., Sivachenko A.Y., Sboner A., Esgueva R., Pflueger D., Sougnez C., et al. . 2011b The genomic complexity of primary human prostate cancer. Nature. 470:214–220. 10.1038/nature09744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F.R., and Ponstingl H.. 1991. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 354:80–82. 10.1038/354080a0 [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network 2015. The molecular taxonomy of primary prostate cancer. Cell. 163:1011–1025. 10.1016/j.cell.2015.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver B.S., Tran J., Gopalan A., Chen Z., Shaikh S., Carracedo A., Alimonti A., Nardella C., Varmeh S., Scardino P.T., et al. . 2009. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat. Genet. 41:619–624. 10.1038/ng.370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano E., and Downward J.. 2010. Role of RAS in the regulation of PI 3-kinase. Curr. Top. Microbiol. Immunol. 346:143–169. 10.1007/82_2010_56 [DOI] [PubMed] [Google Scholar]
- Chen M., Pratt C.P., Zeeman M.E., Schultz N., Taylor B.S., O’Neill A., Castillo-Martin M., Nowak D.G., Naguib A., Grace D.M., et al. . 2011. Identification of PHLPP1 as a tumor suppressor reveals the role of feedback activation in PTEN-mutant prostate cancer progression. Cancer Cell. 20:173–186. 10.1016/j.ccr.2011.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Trotman L.C., Shaffer D., Lin H.K., Dotan Z.A., Niki M., Koutcher J.A., Scher H.I., Ludwig T., Gerald W., et al. . 2005. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 436:725–730. 10.1038/nature03918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Herzka T., Zheng W., Qi J., Wilkinson J.E., Bradner J.E., Robinson B.D., Castillo-Martin M., Cordon-Cardo C., and Trotman L.C.. 2014. RapidCaP, a novel GEM model for analysis and therapy of metastatic prostate cancer, reveals Myc as a driver of Pten-mutant metastasis. Cancer Discov. 4:318-333. 10.1158/2159-8290.CD-13-0346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Herzka T., Stahlhut C., Watrud K., Robinson B.D., and Trotman L.C.. 2015. Rapid in vivo validation of candidate drivers derived from the PTEN-mutant prostate metastasis genome. Methods. 77-78:197–204. 10.1016/j.ymeth.2014.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chook Y.M., and Blobel G.. 2001. Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 11:703–715. 10.1016/S0959-440X(01)00264-0 [DOI] [PubMed] [Google Scholar]
- Christie K.J., Martinez J.A., and Zochodne D.W.. 2012. Disruption of E3 ligase NEDD4 in peripheral neurons interrupts axon outgrowth: Linkage to PTEN. Mol. Cell. Neurosci. 50:179–192. 10.1016/j.mcn.2012.04.006 [DOI] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., and Zhang F.. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science. 339:819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Mitri D., Toso A., Chen J.J., Sarti M., Pinton S., Jost T.R., D’Antuono R., Montani E., Garcia-Escudero R., Guccini I., et al. . 2014. Tumour-infiltrating Gr-1+ myeloid cells antagonize senescence in cancer. Nature. 515:134–137. 10.1038/nature13638 [DOI] [PubMed] [Google Scholar]
- Drinjakovic J., Jung H., Campbell D.S., Strochlic L., Dwivedy A., and Holt C.E.. 2010. E3 ligase Nedd4 promotes axon branching by downregulating PTEN. Neuron. 65:341–357. 10.1016/j.neuron.2010.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A., Andrés-Pons A., Fernández E., Valiente M., Torres J., Cervera J., and Pulido R.. 2006. Nuclear localization of PTEN by a Ran-dependent mechanism enhances apoptosis: Involvement of an N-terminal nuclear localization domain and multiple nuclear exclusion motifs. Mol. Biol. Cell. 17:4002–4013. 10.1091/mbc.E06-05-0380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh C.P., Putz U., Howitt J., Low L.H., Gunnersen J., Bye N., Morganti-Kossmann C., and Tan S.S.. 2014. Nuclear trafficking of Pten after brain injury leads to neuron survival not death. Exp. Neurol. 252:37–46. 10.1016/j.expneurol.2013.11.017 [DOI] [PubMed] [Google Scholar]
- Görlich D., and Kutay U.. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607–660. 10.1146/annurev.cellbio.15.1.607 [DOI] [PubMed] [Google Scholar]
- Grasso C.S., Wu Y.M., Robinson D.R., Cao X., Dhanasekaran S.M., Khan A.P., Quist M.J., Jing X., Lonigro R.J., Brenner J.C., et al. . 2012. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 487:239–243. 10.1038/nature11125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Maccario H., Kriplani N., and Leslie N.R.. 2016. In cell and in vitro assays to measure PTEN ubiquitination. Methods Mol. Biol. 1388:155–165. 10.1007/978-1-4939-3299-3_11 [DOI] [PubMed] [Google Scholar]
- Hamilton M.H., Tcherepanova I., Huibregtse J.M., and McDonnell D.P.. 2001. Nuclear import/export of hRPF1/Nedd4 regulates the ubiquitin-dependent degradation of its nuclear substrates. J. Biol. Chem. 276:26324–26331. 10.1074/jbc.M101205200 [DOI] [PubMed] [Google Scholar]
- Harvey K.F., Shearwin-Whyatt L.M., Fotia A., Parton R.G., and Kumar S.. 2002. N4WBP5, a potential target for ubiquitination by the Nedd4 family of proteins, is a novel Golgi-associated protein. J. Biol. Chem. 277:9307–9317. 10.1074/jbc.M110443200 [DOI] [PubMed] [Google Scholar]
- Hieronymus H., Schultz N., Gopalan A., Carver B.S., Chang M.T., Xiao Y., Heguy A., Huberman K., Bernstein M., Assel M., et al. . 2014. Copy number alteration burden predicts prostate cancer relapse. Proc. Natl. Acad. Sci. USA. 111:11139–11144. 10.1073/pnas.1411446111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander M.C., Blumenthal G.M., and Dennis P.A.. 2011. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat. Rev. Cancer. 11:289–301. 10.1038/nrc3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt J., Lackovic J., Low L.H., Naguib A., Macintyre A., Goh C.P., Callaway J.K., Hammond V., Thomas T., Dixon M., et al. . 2012. Ndfip1 regulates nuclear Pten import in vivo to promote neuronal survival following cerebral ischemia. J. Cell Biol. 196:29–36. 10.1083/jcb.201105009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlader N., Noone A.M., Krapcho M., Miller D., Bishop K., Altekruse S.F., Kosary C.L., Yu M., Ruhl J., and Tatalovich Z., eds. SEER Cancer Statistics Review, 1975-2013, National Cancer Institute. Bethesda, MD. http://seer.cancer.gov/csr/1975_2013/ (based on November 2015 SEER data submission, posted to the SEER web site, April 2016).
- Irshad S., and Abate-Shen C.. 2013. Modeling prostate cancer in mice: Something old, something new, something premalignant, something metastatic. Cancer Metastasis Rev. 32:109–122. 10.1007/s10555-012-9409-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanaga K., Yang Y., Raso M.G., Ma L., Hanna A.E., Thilaganathan N., Moghaddam S., Evans C.M., Li H., Cai W.W., et al. . 2008. Pten inactivation accelerates oncogenic K-ras-initiated tumorigenesis in a mouse model of lung cancer. Cancer Res. 68:1119–1127. 10.1158/0008-5472.CAN-07-3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazgan N., Williams T., Forsberg L.J., and Brenman J.E.. 2010. Identification of a nuclear export signal in the catalytic subunit of AMP-activated protein kinase. Mol. Biol. Cell. 21:3433–3442. 10.1091/mbc.E10-04-0347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T., and Cheeseman I.M.. 2012. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat. Cell Biol. 14:311–317. 10.1038/ncb2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwabi-Addo B., Giri D., Schmidt K., Podsypanina K., Parsons R., Greenberg N., and Ittmann M.. 2001. Haploinsufficiency of the Pten tumor suppressor gene promotes prostate cancer progression. Proc. Natl. Acad. Sci. USA. 98:11563–11568. 10.1073/pnas.201167798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie N.R., and Foti M.. 2011. Non-genomic loss of PTEN function in cancer: not in my genes. Trends Pharmacol. Sci. 32:131–140. 10.1016/j.tips.2010.12.005 [DOI] [PubMed] [Google Scholar]
- Li J., Yen C., Liaw D., Podsypanina K., Bose S., Wang S.I., Puc J., Miliaresis C., Rodgers L., McCombie R., et al. . 1997. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 275:1943–1947. 10.1126/science.275.5308.1943 [DOI] [PubMed] [Google Scholar]
- Liu W., Laitinen S., Khan S., Vihinen M., Kowalski J., Yu G., Chen L., Ewing C.M., Eisenberger M.A., Carducci M.A., et al. . 2009. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat. Med. 15:559–565. 10.1038/nm.1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsit C.J., Zheng S., Aldape K., Hinds P.W., Nelson H.H., Wiencke J.K., and Kelsey K.T.. 2005. PTEN expression in non-small-cell lung cancer: evaluating its relation to tumor characteristics, allelic loss, and epigenetic alteration. Hum. Pathol. 36:768–776. 10.1016/j.humpath.2005.05.006 [DOI] [PubMed] [Google Scholar]
- Melchior F., Paschal B., Evans J., and Gerace L.. 1993. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J. Cell Biol. 123:1649–1659. 10.1083/jcb.123.6.1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguib A., and Trotman L.C.. 2013. PTEN plasticity: How the taming of a lethal gene can go too far. Trends Cell Biol. 23:374–379. 10.1016/j.tcb.2013.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardella C., Clohessy J.G., Alimonti A., and Pandolfi P.P.. 2011. Pro-senescence therapy for cancer treatment. Nat. Rev. Cancer. 11:503–511. 10.1038/nrc3057 [DOI] [PubMed] [Google Scholar]
- Nowak D.G., Cho H., Herzka T., Watrud K., DeMarco D.V., Wang V.M., Senturk S., Fellmann C., Ding D., Beinortas T., et al. . 2015. MYC drives Pten/Trp53-deficient proliferation and metastasis due to IL6 secretion and AKT suppression via PHLPP2. Cancer Discov. 5:636-651. 10.1158/2159-8290.CD-14-1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plafker S.M., and Macara I.G.. 2000. Importin-11, a nuclear import receptor for the ubiquitin-conjugating enzyme, UbcM2. EMBO J. 19:5502–5513. 10.1093/emboj/19.20.5502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plafker S.M., Plafker K.S., Weissman A.M., and Macara I.G.. 2004. Ubiquitin charging of human class III ubiquitin-conjugating enzymes triggers their nuclear import. J. Cell Biol. 167:649–659. 10.1083/jcb.200406001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podsypanina K., Ellenson L.H., Nemes A., Gu J., Tamura M., Yamada K.M., Cordon-Cardo C., Catoretti G., Fisher P.E., and Parsons R.. 1999. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc. Natl. Acad. Sci. USA. 96:1563–1568. 10.1073/pnas.96.4.1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach M., and Blobel G.. 1995. Protein import into nuclei: Association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 83:683–692. 10.1016/0092-8674(95)90181-7 [DOI] [PubMed] [Google Scholar]
- Robinson D., Van Allen E.M., Wu Y.M., Schultz N., Lonigro R.J., Mosquera J.M., Montgomery B., Taplin M.E., Pritchard C.C., Attard G., et al. . 2015. Integrative clinical genomics of advanced prostate cancer. Cell. 161:1215–1228. (published erratum appears in Cell. 2015. 162:454) 10.1016/j.cell.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrima M., De Marco C., Fabiani F., Franco R., Pirozzi G., Rocco G., Ravo M., Weisz A., Zoppoli P., Ceccarelli M., et al. . 2012. Signaling networks associated with AKT activation in non-small cell lung cancer (NSCLC): New insights on the role of phosphatydil-inositol-3 kinase. PLoS One. 7:e30427 10.1371/journal.pone.0030427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M.S., Salmena L., and Pandolfi P.P.. 2012. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 13:283–296. 10.1038/nrm3330 [DOI] [PubMed] [Google Scholar]
- Taniguchi C.M., Winnay J., Kondo T., Bronson R.T., Guimaraes A.R., Alemán J.O., Luo J., Stephanopoulos G., Weissleder R., Cantley L.C., and Kahn C.R.. 2010. The phosphoinositide 3-kinase regulatory subunit p85alpha can exert tumor suppressor properties through negative regulation of growth factor signaling. Cancer Res. 70:5305–5315. 10.1158/0008-5472.CAN-09-3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B.S., Schultz N., Hieronymus H., Gopalan A., Xiao Y., Carver B.S., Arora V.K., Kaushik P., Cerami E., Reva B., et al. . 2010. Integrative genomic profiling of human prostate cancer. Cancer Cell. 18:11–22. 10.1016/j.ccr.2010.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman L.C., Niki M., Dotan Z.A., Koutcher J.A., Di Cristofano A., Xiao A., Khoo A.S., Roy-Burman P., Greenberg N.M., Van Dyke T., et al. . 2003. Pten dose dictates cancer progression in the prostate. PLoS Biol. 1:E59 10.1371/journal.pbio.0000059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman L.C., Wang X., Alimonti A., Chen Z., Teruya-Feldstein J., Yang H., Pavletich N.P., Carver B.S., Cordon-Cardo C., Erdjument-Bromage H., et al. . 2007. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 128:141–156. 10.1016/j.cell.2006.11.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B., Stephens L., and Hawkins P.. 2012. PI3K signalling: The path to discovery and understanding. Nat. Rev. Mol. Cell Biol. 13:195–203. 10.1038/nrm3290 [DOI] [PubMed] [Google Scholar]
- Wagner P.L., Stiedl A.C., Wilbertz T., Petersen K., Scheble V., Menon R., Reischl M., Mikut R., Rubin M.A., Fend F., et al. . 2011. Frequency and clinicopathologic correlates of KRAS amplification in non-small cell lung carcinoma. Lung Cancer. 74:118–123. 10.1016/j.lungcan.2011.01.029 [DOI] [PubMed] [Google Scholar]
- Wang S.I., Parsons R., and Ittmann M.. 1998. Homozygous deletion of the PTEN tumor suppressor gene in a subset of prostate adenocarcinomas. Clin. Cancer Res. 4:811–815. [PubMed] [Google Scholar]
- Wang X., Trotman L.C., Koppie T., Alimonti A., Chen Z., Gao Z., Wang J., Erdjument-Bromage H., Tempst P., Cordon-Cardo C., et al. . 2007. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 128:129–139. 10.1016/j.cell.2006.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtalla A., and Arcaro A.. 2011. Targeting phosphoinositide 3-kinase signalling in lung cancer. Crit. Rev. Oncol. Hematol. 80:278–290. 10.1016/j.critrevonc.2011.01.007 [DOI] [PubMed] [Google Scholar]
- Yanagawa N., Leduc C., Kohler D., Saieg M.A., John T., Sykes J., Yoshimoto M., Pintilie M., Squire J., Shepherd F.A., and Tsao M.S.. 2012. Loss of phosphatase and tensin homolog protein expression is an independent poor prognostic marker in lung adenocarcinoma. J. Thorac. Oncol. 7:1513–1521. 10.1097/JTO.0b013e3182641d4f [DOI] [PubMed] [Google Scholar]
- Yanagi S., Kishimoto H., Kawahara K., Sasaki T., Sasaki M., Nishio M., Yajima N., Hamada K., Horie Y., Kubo H., et al. . 2007. Pten controls lung morphogenesis, bronchioalveolar stem cells, and onset of lung adenocarcinomas in mice. J. Clin. Invest. 117:2929–2940. 10.1172/JCI31854 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.