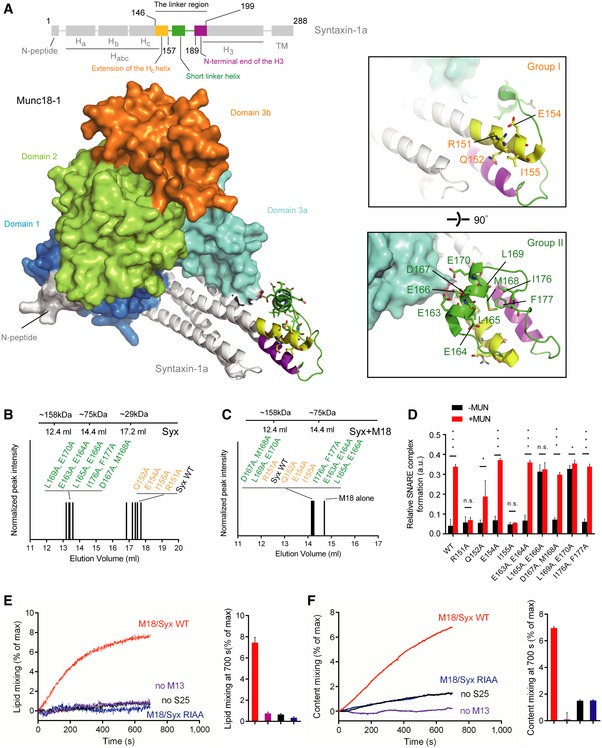
Figure 1. Identification of the key residues in syntaxin‐1 that are essential for Munc13‐1 activity in ternary SNARE complex formation.
-
ADomain structure of syntaxin‐1 (top) and crystal structure of the closed Munc18‐1/syntaxin‐1 complex with annotations (bottom, PDB ID 3C98). The insets show two close‐up views of the syntaxin‐1 linker region. Labels correspond to residues that were mutated.
-
B, CSize‐exclusion chromatography peak elution volumes of mutants of the syntaxin‐1 linker region in the absence (B) and presence of Munc18‐1 (C) using a 24 ml Superdex 200 column.
-
DRelative efficiencies of ternary SNARE complex formation using syntaxin‐1 with selected mutations in the linker region. The reactions were performed in the presence of Munc18‐1 and/or the MUN domain and monitored by ensemble FRET efficiency between fluorescent dye‐labeled SNAP‐25 and synaptobrevin‐2. Shown are means ± SD; n.s., not significant (P > 0.05); *P < 0.05; ***P < 0.001, using two‐tailed Student's t‐test with n = 5 technical replicates.
-
E, FEnsemble lipid (E) and content mixing (F) between proteoliposomes with reconstituted Munc18‐1/syntaxin‐1 complex and proteoliposomes with reconstituted synaptobrevin‐2 and synaptotagmin‐1 in the presence of 1 mM Ca2+. Bar charts are means ± SD for n = 3 technical replicates. M18, Munc18‐1; M13, Munc13‐1 C1‐C2B‐MUN domain; S25, SNAP‐25; Syx, syntaxin‐1.
