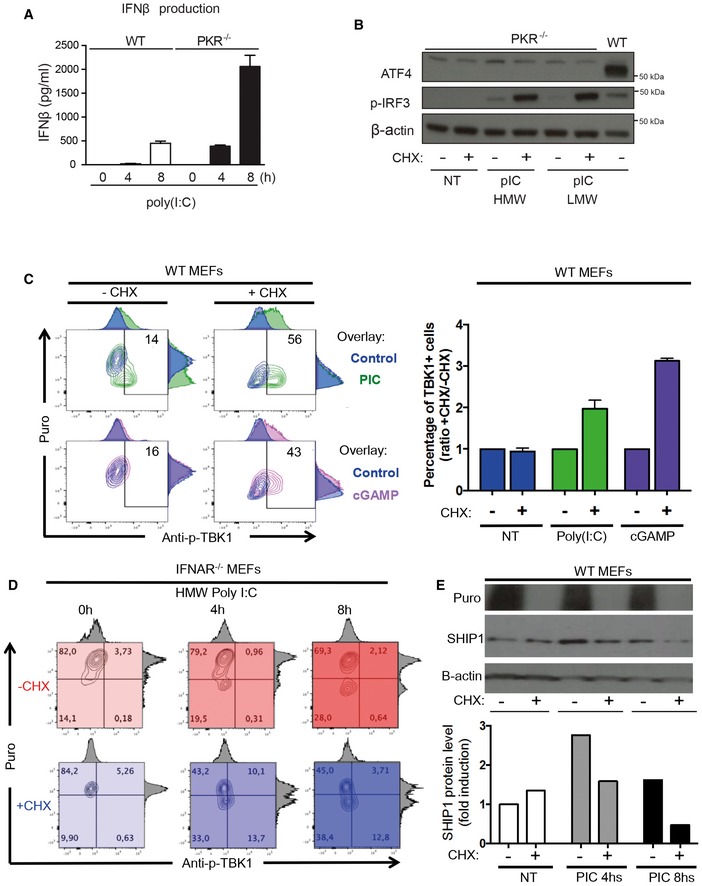
IFN‐β production was measured by ELISA in cell culture supernatants of WT and PKR−/− MEFs that were stimulated with HMW poly(I:C) for 4 and 8 h. Results are the mean ± SD of three independent experiments.
PKR−/− MEFs were left untreated (NT) or were treated with HMW or LMW poly(I:C) for 8 h in the presence or not of 5 μg/ml of cycloheximide (CHX). ATF4 and phospho‐IRF3 levels in nuclear extracts were detected by immunoblot. β‐actin was used as equal loading control. WT MEFs were treated with LMW poly(I:C) for 4 h and the corresponding nuclear extract was used as a positive control for ATF4 detection.
WT MEFs were lipofected with poly(I:C) (green) or cGAMP (pink) for the indicated time in the presence or absence of CHX. Cells were pulsed with puromycin prior to collection, staining with anti‐puromycin and anti‐pTBK1 and flow cytometry analysis. Quantification is presented as histograms on the right (n ≥ 2). Bars are mean ± SD.
IFNAR1−/− MEFs were lipofected with poly(I:C) for the indicated time in the presence (shades of blue) or absence (shades of red) of CHX. Cells were pulsed with puromycin prior to collection, staining with anti‐puromycin and anti‐pTBK1, and flow cytometry analysis.
IFNAR1−/− MEFs were lipofected with poly(I:C) for the indicated time in the presence or absence of CHX. Cells were pulsed with puromycin prior to collection and immunoblot detection of puromycin and SHIP‐1. β‐actin was used as equal loading control. Quantification of the blot is presented below.