Figure 2.
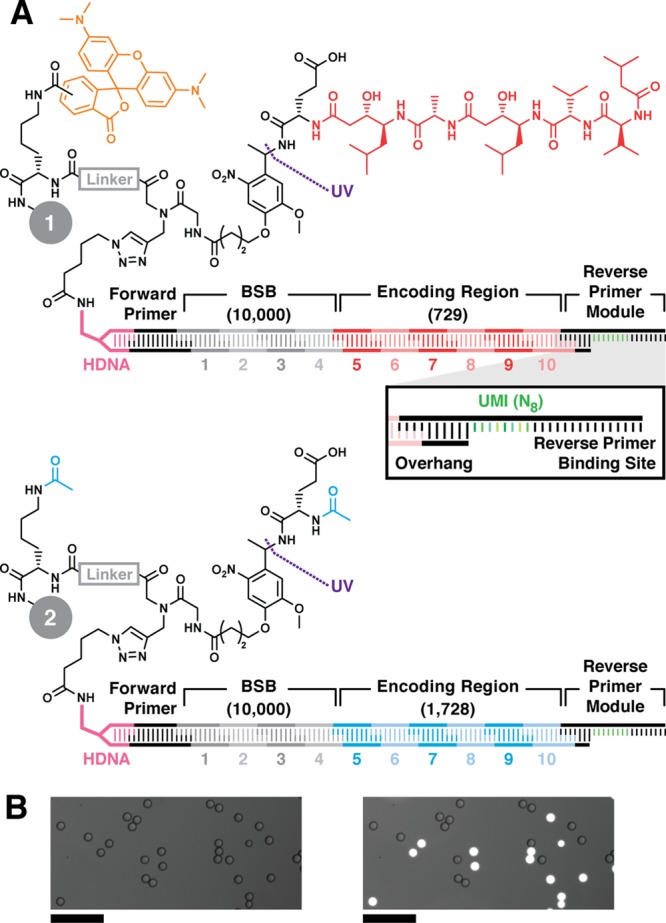
Model DNA-encoded library bead structures. Lysine, linker (gray), photocleavable linker, and Glu were sequentially coupled to 10-μm-diameter TentaGel resin. (A) Positive control inhibitor beads 1 display pepstatin A (red) coupled to Glu. The linker is labeled with 5(6)-carboxy TMR fluorophore (orange). Negative control beads 2 were prepared by acetylating the Glu α amine and linker amine (cyan). Bead sets were substoichiometrically functionalized with azido DNA headpiece (HDNA) via CuAAC. The DNA encoding sequence was installed by split-and-pool combinatorial enzymatic ligation. The BSB region contained 10 unique sequence modules at each of 4 positions (1—4, 104 possible BSBs). The encoding regions (ER, 5—10) contained either 729 (Glu-pepstatin A positive control beads, 1) or 1728 (N-acetyl-Glu negative control beads, 2) possible sequences. The DNA sequence terminates with ligation of a reverse primer module containing the reverse PCR primer binding site flanking an internal unique molecular identifier (UMI, green, inset). The UMI is a random 8-mer (65 536 possible sequences). (B) Micrographs of a model library containing positive and negative control beads 1 and 2 visualized in brightfield (left) and brightfield overlay with TMR fluorescence emission (λex = 550 nm; λem = 570 nm; right) illustrate facile differentiation between the two bead types. Scale = 100 μm.
