Abstract
Emerging evidence suggests that DEAH-box polypeptide 32 (DHX32) serves an important role in the progression and metastasis of cancer. However, the role of DHX32 in breast cancer remains to be completely elucidated. The aim of the present study was to evaluate the expression and clinical significance of DHX32 in breast cancer. The reverse transcription-quantitative polymerase chain reaction was performed to analyze DHX32 messenger (m)RNA expression, and western blotting and immunohistochemistry were performed to examine DHX32 protein expression in breast cancer and adjacent non-cancerous tissues. The association in breast cancer between DHX32 expression, clinicopathological features and prognosis was analyzed using 193 breast cancer tissue samples. The results of the present study demonstrated that breast cancer tissues exhibited increased DHX32 mRNA and protein expression compared with adjacent non-cancerous tissues (P<0.001). In addition, DHX32 expression was significantly associated with breast cancer clinical stage (P=0.006), histological grade (P=0.029), lymph node metastasis (P<0.001) and expression of the proliferation marker Ki-67 (P=0.004). Kaplan-Meier estimator analysis indicated that increased DHX32 expression is associated with poor prognosis in patients with breast cancer. Furthermore, the Cox proportional hazards model indicated that DHX32 expression is an independent prognostic factor for decreased overall survival and disease-free survival in patients with breast cancer. In conclusion, the results of the present study suggest that DHX32 overexpression is an unfavorable prognostic biomarker in breast cancer and a potential therapeutic target of future breast cancer treatments.
Keywords: breast cancer, DEAH-box helicase 32, prognosis, biomarker
Introduction
Breast cancer is the most frequently diagnosed cancer in women, with 425,000 new cases reported each year and an annual mortality of 78,000 individuals in China (1). Despite the existence of a variety of breast cancer treatments, including surgical resection, adjuvant chemotherapy, radiotherapy, hormone therapy and targeted therapy, breast cancer remains the second leading cause of cancer-associated mortality in women worldwide (2). To date, a number of molecules have been demonstrated to serve roles in breast cancer progression and metastasis, and the discovery of biomarkers, including HER2, has led to targeted treatment options (3); however, the underlying mechanisms of breast cancer development and progression remain unclear. Therefore, the identification of novel breast cancer biomarkers is required to improve the determination of cancer prognosis and the development of treatments.
Human RNA helicases constitute an extended family of enzymes that serve important roles in numerous aspects of RNA metabolism, including splicing, transcription, translation and degradation (4,5). DEAH-box polypeptide 32 (DHX32) is a novel RNA helicase containing a unique helicase domain structure and has been observed to be dysregulated in certain types of tumor (6,7). In addition, a number of other RNA helicases have been demonstrated to be dysregulated in various types of cancer, although their exact role in carcinogenesis remains to be completely elucidated (8,9). RNA helicases are involved in the regulation of multiple cancer-associated molecules, including DEAD-box helicase 5 (DDX5) (10), DDX17 (11) and DHX9 (12–14). Human DHX32 exhibits widespread tissue distribution, including colon, breast and lung tissue. In addition, the amino acid sequence of human DHX32 is highly homologous to that of its murine counterpart, with 84% identity and 90% similarity, indicating that it is an evolutionally conserved and functionally important gene (15). A previous study demonstrated that DHX32 is overexpressed in colorectal cancer (CRC) tissue samples compared with adjacent wild-type tissue (7). In addition, DHX32 expression is significantly associated with clinicopathological features of CRC, which suggests that DHX32 may be used as a prognostic biomarker in patients with CRC (7). A recent study observed that DHX32 promotes the proliferation, migration and invasion of CRC cells by activating the Wnt signaling pathway and downregulating pro-apoptotic gene expression (16). This suggests that DHX32 expression is associated with CRC development and progression (16). However, the exact role of DHX32 in breast cancer remains unclear.
In the present study, the expression of DHX32 in breast cancer and adjacent non-cancerous tissue samples was investigated, demonstrating that DHX32 was markedly upregulated in human breast cancer specimens. Furthermore, the association between DHX32 expression and the clinicopathological features of patients with breast cancer was examined to assess whether DHX32 is a potential prognostic indicator in breast cancer.
Materials and methods
Patients and specimens
A total of 193 paraffin-embedded breast cancer specimens were obtained from the Beijing Tiantan Hospital (Beijing, China) between June 2007 and December 2009. In addition, a total of 40 pairs of freshly frozen breast cancer tissue samples and adjacent normal mammary tissue samples were collected during surgery at the Beijing Tiantan Hospital between May 2014 and September 2014. All fresh samples were snap-frozen in liquid nitrogen and stored at −80°C until required and none of the patients received chemotherapy or radiotherapy prior to surgery. Breast cancer diagnosis was confirmed by histological analysis of tissue sections stained with hematoxylin and eosin, and complete clinical data of the 193 breast cancer cases was obtained and reviewed. Male breast cancer patients were excluded from the present study. The clinical follow-up time was 60 months. The overall survival time (OS) was calculated as the time between the date of surgery and breast cancer-associated mortality. The disease-free survival time (DFS) was calculated as the time between the date of surgery and initial tumor progression, which was indicative of initial recurrence of cancer. Table I presents the clinicopathological characteristics of the 193 patients. The present study was approved by the Ethics Committee of Beijing Tiantan Hospital (Beijing, China) and written informed consent was obtained from all subjects.
Table I.
Clinicopathological characteristics of 193 patients with breast cancer.
| Characteristic | Number of patients (%) |
|---|---|
| Age (years) | |
| ≤50 | 87 (45.1) |
| ≥50 | 106 (54.9) |
| Tumor size (cm) | |
| ≤2 | 120 (62.2) |
| ≥2 | 73 (37.8) |
| Histological grade | |
| I–II | 114 (59.1) |
| III | 79 (40.9) |
| Clinical stage | |
| I–II | 128 (66.3) |
| III–IV | 65 (33.7) |
| Lymph node metastasis | |
| Negative | 117 (60.6) |
| Positive | 76 (39.4) |
| Estrogen receptor status | |
| Negativeb | 85 (44.0) |
| Positivec | 108 (56.0) |
| Progesterone receptor status | |
| Negativeb | 74 (38.3) |
| Positivec | 119 (61.7) |
| HER2 status | |
| Negativeb | 136 (70.5) |
| Positivec | 57 (29.5) |
| Ki-67 expression | |
| Negativeb | 60 (31.1) |
| Positivec | 133 (68.9) |
| Expression of DHX32a | |
| Low | 89 (46.1) |
| High | 104 (53.9) |
High or low DHX32 expression was determined by immunohistochemistry analysis.
Absent or weak staining was categorized as negative.
Moderate or strong staining was categorized as positive. HER2, human epidermal growth factor receptor-2; Ki-67, marker of proliferation Ki-67; DHX32, DEAH-box helicase 32.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
DHX32 messenger (m)RNA expression in 40 pairs of breast cancer and adjacent non-cancerous tissues was detected using RT-qPCR. Total RNA was extracted using TRIzol® reagent (Invitrogen; Thermo fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol. RT-qPCR was performed using a Thermal Cycler Dice Real Time System TP800 (Takara Bio, Inc., Otsu, Japan) and the SYBR Premix ExTaq kit (Takara Biotechnology Co., Ltd., Dalian, China) to detect DHX32 mRNA expression, according to the manufacturer's protocol. GAPDH was used as an internal control. The PCR primers for DHX32 and GAPDH are shown in Table II. The thermocycling conditions for RT-qPCR were as follows: 95°C for 30 sec; and 40 cycles of 95°C for 5 sec, 51°C for 20 sec and 72°C for 30 sec. Each reaction was performed in triplicate, and the mean DHX32 mRNA level for each tumor sample was compared with its adjacent non-cancerous tissue sample. The relative expression of DHX32 was calculated using the 2−ΔΔCq method (17) and normalized to GAPDH.
Table II.
Sequences of primers used in reverse transcription- quantitative polymerase chain reaction analysis.
| Primer | Sequence |
|---|---|
| DHX32 | |
| Forward | 5′-GCCTGTGAGGATTTGGAACT-3′ |
| Reverse | 5′-GCTGGTAGTGGATGGAAAGACA-3′ |
| GAPDH | |
| Forward | 5′-CTATAAATTGAGCCCGCAGCC-3′ |
| Reverse | 5′-GCGCCCAATACGACCAAATC-3′ |
DHX32, DEAH-box helicase 32.
Western blot analysis
A total of eight randomly selected fresh tissue samples were lysed in protein lysis buffer containing 50 mmol/l Tris-HCl (pH 7.4), 150 mmol/l NaCl, 1% NP-40, 0.1% SDS and a protease inhibitor cocktail, followed by centrifugation (12,000 × g for 20 min at 4°C). Protein concentrations were determined using a bicinchoninic acid protein quantitation kit (Applygen Technologies, Inc., Beijing, China). A total of 50 µg each protein sample was separated by SDS-PAGE on a 12% gel and transferred to a polyvinylidene fluoride membrane. Following blocking in PBS with Tween 20 add 5% skim milk powder for 1 h at room temperature, the membrane was incubated with anti-DHX32 rabbit antibody (catalog no. 19808-1-AP; dilution, 1:500; Proteintech Group, Inc., Chicago, IL, USA) at 4°C overnight. Following three washes in TBS plus Tween 20 buffer, the membranes were incubated with a horseradish peroxidase (HRP)-conjugated anti-rabbit immunoglobulin G (IgG) secondary antibody (catalog no., HS101-01; dilution, 1:2,000; Beijing TransGen Biotech Co., Ltd., Beijing, China) for 1 h at room temperature. Immunoreactive protein bands were visualized using an enhanced chemiluminescence reagent (Applygen Technologies, Inc.). β-actin was used as a loading control, and the membrane was incubated with anti-β-actin mouse monoclonal antibody (catalog no. HC201-01; dilution, 1:2,000; Beijing TransGen Biotech Co., Ltd.) at 4°C overnight. Upon washing, the membranes were incubated with an HRP-conjugated anti-mouse IgG secondary antibody (catalog no. HS201-01; dilution, 1:2,000; Beijing TransGen Biotech Co., Ltd.) for 1 h at room temperature. Other steps were the same as above.
Immunohistochemistry (IHC) analysis
Paraffin-embedded sections from 193 clinical specimens were deparaffinized in xylene and rehydrated in a descending ethanol series. Heat-induced antigen retrieval was performed in 0.01 M citrate buffer (pH 6.0) at 100°C for 10 min. Endogenous peroxidase activity was inhibited by immersion in 0.3% hydrogen peroxide for 10 min. To block the sections, 5% bovine serum albumin (Beijing Solarbio Science & Technology Co., Ltd, Beijing, China) in PBS was used for 1 h at room temperature. The sections were next incubated with an anti-DHX32 rabbit monoclonal antibody (catalog no. 19808-1-AP; dilution, 1:100; Proteintech Group, Inc.) overnight at 4°C. Following washing with PBS, sections were incubated with an HRP-labeled IgG secondary antibody (catalog no., KIHC-1; Proteintech Group, Inc.) for 30 min at room temperature. The immunoreactive products were visualized using the 3,3′-Diaminobenzidine Color Development kit (ZSGB Biotech, Beijing, China). Following washing in water, sections were counterstained with hematoxylin and dehydrated in an ascending series of methanol, prior to being cleaned in xylene and mounted on a coverslip. Negative controls were processed in parallel following the same protocol, but substituting the primary antibody for PBS.
The stained tissue sections were examined by an evaluator who was unaware of the clinicopathological features of each sample. DHX32 protein expression was classified semi-quantitatively according to the synthetic evaluation of the quantity and intensity of positively stained cells, as described below. The percentage of positively stained cells was grouped into four grades: 0 points (0%), 1 point (1–10%), 2 points (10–50%) and 3 points (>50%). Staining intensity was segmented into four ranks: 0 points (no staining), 1 point (weak staining), 2 points (moderate staining) and 3 points (strong staining). The status of DHX32 expression was determined by the total points from percentage of positive cells and staining intensity, as low expression (0–3 points) or high expression (4–6 points).
Statistical analysis
All data were analyzed using SPSS software version 20.0 (IBM SPSS, Armonk, NY, USA). RT-qPCR results are presented as the mean ± standard deviation. The difference in DHX32 mRNA expression between breast cancer and adjacent non-cancerous tissue samples was assessed using the Wilcoxon signed-rank test. Pearson's χ2 test and Fisher's exact test were used to analyze the association between DHX32 expression and clinicopathological features of breast cancer. Survival data were evaluated using the Kaplan-Meier estimator and log-rank test. Multivariate analysis was performed using the Cox proportional hazards model. Univariate and multivariate Cox proportional hazards regression analyses were performed to evaluate the impact of DHX32 expression and other clinicopathological factors on OS and DFS. P<0.05 was considered to indicate a statistically significant difference.
Results
DHX32 expression is increased in breast cancer
To identify the role of DHX32 expression in breast cancer, RT-qPCR was performed to evaluate the mRNA expression in 40 pairs of breast cancer and adjacent normal mammary tissues, with GAPDH serving as the internal control. Compared with adjacent non-cancerous tissue, breast cancer tissue exhibited increased DHX32 mRNA expression (P<0.001; Fig. 1). Furthermore, the DHX32 protein expression of eight randomly selected breast cancer and adjacent non-cancerous tissue samples was evaluated using western blotting. DHX32 was observed to be overexpressed in the breast cancer tissue samples compared with the non-cancerous tissue samples (Fig. 2). These results were consistent with the IHC analyses, which demonstrated increased DHX32 expression in the breast cancer tissue samples, localized primarily to the cytoplasm, compared with the adjacent non-cancerous tissue samples (Fig. 3).
Figure 1.
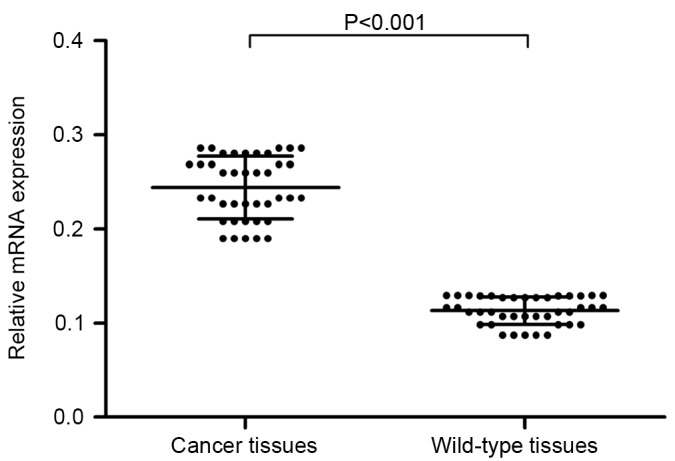
Reverse transcription-quantitative polymerase chain reaction analysis demonstrated significantly increased DHX32 mRNA expression in the breast cancer tissue samples compared with that in the adjacent wild-type mammary tissues (P<0.001, Wilcoxon matched-pairs signed-ranks test). DHX32, DEAH-box helicase 32; mRNA, messenger RNA.
Figure 2.
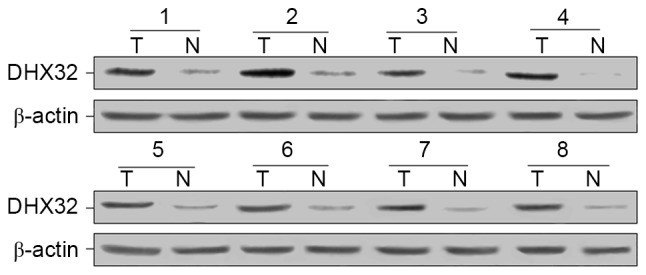
Western blot analysis demonstrated increased DHX32 protein expression in the breast cancer tissue samples compared with the adjacent wild-type mammary tissue samples. Numbers between 1 and 8 represent pairs of tissue samples. DHX32, DEAH-box helicase 32; T, tumor tissue sample; N, normal, adjacent wild-type tissue sample.
Figure 3.
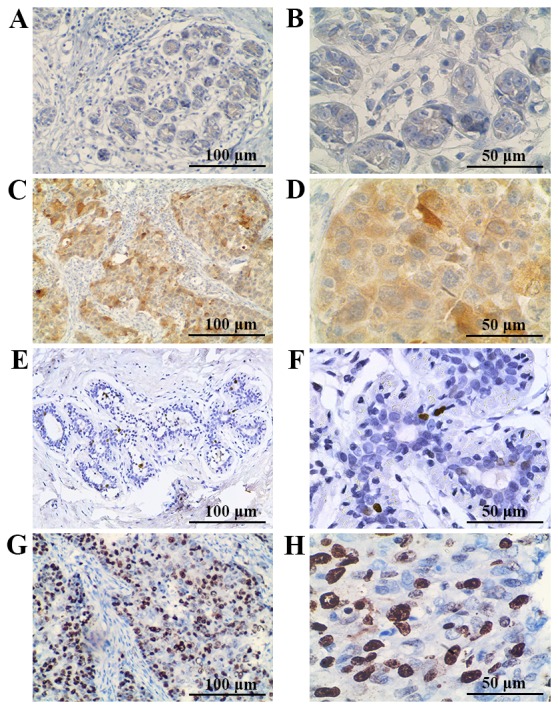
Immunohistochemical staining of DHX32 and Ki-67 proteins in breast tissue samples. DHX32 staining of adjacent non-cancerous mammary tissue at a magnification of (A) ×200 and (B) ×400. DHX32 staining of breast cancer tissue at a magnification of (C) ×200 and (D) ×400. Ki-67 staining of adjacent non-cancerous mammary tissue at a magnification of (E) ×200 and (F) ×400. Ki-67 staining of breast cancer tissue at a magnification of (G) ×200 and (H) ×400. DHX32, DEAH-box helicase 32; Ki-67, marker of proliferation Ki-67.
Association between DHX32 protein expression and the clinicopathological characteristics of patients with breast cancer
DHX32 protein expression was assessed in 193 archived paraffin-embedded breast cancer samples using IHC staining. The 193 breast cancer patients were divided into two groups: A low DHX32 expression group and a high DHX32 expression group. DHX32 protein expression was then compared with the clinicopathological characteristics of the patients. The clinicopathological characteristics of all of the subjects are indicated in Table I. As summarized in Table III, high expression of DHX32 was significantly associated with an increased clinical stage (I–II vs. III–IV; P=0.006) and histological grade (I–II vs. III; P=0.029), and a positive lymph node metastasis (positive vs. negative; P<0.001) and Ki-67 expression (positive vs. negative; P=0.004) status of the tumor, but was not associated with other clinicopathological parameters.
Table III.
Correlation between DHX32 protein expression and clinicopathological characteristics of patients with breast cancer.
| Characteristic | Low DHX32 expressiona | High DHX32 expressiona | χ2 | P-value |
|---|---|---|---|---|
| Age (years) | 0.539 | |||
| ≤50 | 38 | 49 | 0.378 | |
| ≥50 | 51 | 55 | ||
| Tumor size (cm) | 0.275 | |||
| ≤2 | 59 | 61 | 1.190 | |
| ≥2 | 30 | 43 | ||
| Histological grade | 0.029b | |||
| I–II | 60 | 54 | 4.761 | |
| III | 29 | 50 | ||
| Clinical stage | 0.006c | |||
| I–II | 68 | 60 | 7.518 | |
| III–IV | 21 | 44 | ||
| Lymph node metastasis | <0.001c | |||
| Negative | 69 | 48 | 19.776 | |
| Positive | 20 | 56 | ||
| Estrogen receptor status | 0.815 | |||
| Negatived | 40 | 45 | 0.055 | |
| Positivee | 49 | 59 | ||
| Progesterone receptor status | 0.795 | |||
| Negatived | 35 | 39 | 0.068 | |
| Positivee | 54 | 65 | ||
| HER-2 status | 0.344 | |||
| Negatived | 65 | 71 | 0.897 | |
| Positivee | 23 | 34 | ||
| Ki-67 expression | 0.004c | |||
| Negatived | 37 | 23 | 8.475 | |
| Positivee | 52 | 81 |
High or low DHX32 expression was determined by immunohistochemistry analysis.
P<0.05
P<0.01, as calculated using the Pearson's χ2 and Fisher's exact tests.
Absent or weak staining was categorized as negative.
Moderate or strong staining was categorized as positive. DHX32, DEAH-box helicase 32; HER2, human epidermal growth factor receptor-2; Ki-67, marker of proliferation Ki-67.
Prognostic significance of DHX32 expression in breast cancer
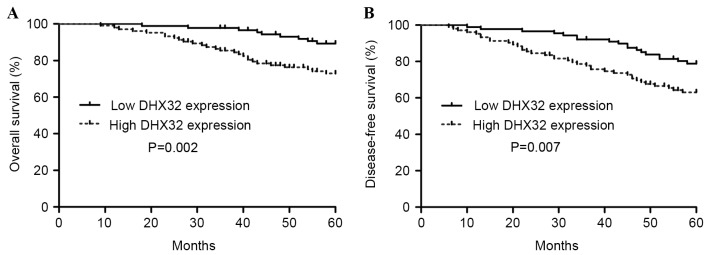
To investigate the prognostic significance of DHX32 in breast cancer, the effect of DHX32 protein expression on OS and DFS in 193 samples from breast cancer patients was assessed using the Kaplan-Meier estimator and the log-rank test. DHX32 protein expression was observed to be associated with OS and DFS in patients with breast cancer (Fig. 4). Patients with a high expression of DHX32 exhibited significantly decreased OS (P=0.002) and DFS (P=0.007) compared with patients with a low DHX32 expression. To evaluate the impact of each variable on OS and DFS, univariate and multivariate Cox proportional hazard regression analyses were used to evaluate the impact of breast cancer DHX32 protein expression and clinicopathological parameters on the prognosis of patients (Table IV). Univariate analysis indicated that the significant factors associated with OS and DFS included clinical stage, histological grade, lymph node metastasis, erb-b2 receptor tyrosine kinase 2 (HER-2) status, Ki-67 expression and DHX32 expression (all P<0.05; exact P-values shown in Table IV). The multivariate analyses indicated that DHX32 protein expression, clinical stage, lymph node metastasis and HER-2 status are significant independent prognostic factors for the OS and DFS of patients with breast cancer (all P<0.05; exact P-values shown in Table IV).
Figure 4.
Kaplan-Meier estimator curves for the correlation between overall survival or disease-free survival and DHX32 protein expression. (A) Overall survival curves of patients with breast cancer. P=0.002 for high vs. low DHX23 expression, as calculated by the log-rank test. (B) Disease-free survival curves of patients with breast cancer. P=0.007 for high vs. low DHX23 expression, as calculated by the log-rank test. High or low DHX32 expression was determined by immunohistochemistry analysis. DHX32, DEAH-box helicase 32.
Table IV.
Univariate and multivariate Cox proportional hazard regression analysis of overall survival and disease-free survival in patients with breast cancer.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Overall survival | ||||||
| Age | 0.734 | 0.388–1.388 | 0.342 | |||
| Tumor size | 1.388 | 0.734–2.624 | 0.313 | |||
| Histological grade | 2.002 | 1.055–3.797 | 0.034a | 1.416 | 0.746–2.689 | 0.288 |
| Clinical stage | 2.500 | 1.322–4.729 | 0.005b | 2.405 | 1.187–4.872 | 0.015a |
| Lymph node metastasis | 3.715 | 1.899–7.266 | <0.001b | 2.809 | 1.398–5.644 | 0.004b |
| Estrogen receptor status | 1.288 | 0.672–2.469 | 0.446 | |||
| Progesterone receptor status | 1.587 | 0.787–3.200 | 0.196 | |||
| HER-2 status | 2.255 | 1.168–4.352 | 0.015a | 2.086 | 1.032–4.214 | 0.041a |
| Ki-67 expression | 2.256 | 1.059–4.807 | 0.035a | 1.512 | 0.658–3.478 | 0.330 |
| DHX32 Expression | 3.144 | 1.335–7.405 | 0.009b | 3.297 | 1.213–8.961 | 0.019a |
| Disease-free survival | ||||||
| Age | 0.787 | 0.439–1.412 | 0.422 | |||
| Tumor size | 1.380 | 0.768–2.479 | 0.281 | |||
| Histological grade | 1.824 | 1.016–3.276 | 0.044a | 1.360 | 0.676–2.737 | 0.389 |
| Clinical stage | 2.427 | 1.254–4.698 | 0.008b | 2.151 | 1.116–4.143 | 0.022a |
| Lymph node metastasis | 3.830 | 2.058–7.125 | <0.001b | 2.712 | 1.478–4.976 | 0.001b |
| Estrogen receptor status | 1.303 | 0.713–2.381 | 0.390 | |||
| Progesterone receptor status | 1.601 | 0.840–3.050 | 0.153 | |||
| HER-2 status | 2.346 | 1.310–4.202 | 0.004b | 2.055 | 1.130–3.738 | 0.018a |
| Ki-67 expression | 2.588 | 0.794–3.096 | 0.024a | 1.535 | 0.714–3.298 | 0.273 |
| DHX32 expression | 2.210 | 1.188–4.110 | 0.012a | 2.210 | 1.073–4.551 | 0.031a |
P<0.05
P<0.01, as calculated using univariate and multivariate Cox proportional hazard regression analyses. HER2, human epidermal growth factor receptor-2; Ki-67, marker of proliferation Ki-67; DHX32, DEAH-box helicase 32; HR, hazard ratio; CI, confidence interval.
Discussion
Breast cancer is one of the most common cancers in women worldwide. Although progress has been made in treatment options for this disease, the morbidity and mortality rates associated with breast cancer continue to increase in various countries in South America, Africa and Asia (18). Therefore, in addition to the established molecular markers, there is a requirement to identify novel biomarkers and therapeutic targets. In the present study, DHX32, a novel RNA helicase, was observed to be overexpressed in breast cancer tissue samples compared with adjacent non-cancerous tissue samples. In addition, the results of the present study indicated that increased DHX32 expression in breast cancer is significantly associated with an increased clinical stage and histological grade, and a positive lymph node metastasis and Ki-67 expression status of the tumor. Furthermore, increased DHX32 expression in breast cancer cells was significantly associated with decreased OS and FDS of the patients. These results indicate that increased DHX32 expression is a prognostic marker in breast cancer.
DHX32 was identified to be a novel RNA helicase containing a unique helicase domain, though it shares similarity with the DHX family of RNA helicases (4). RNA helicases are evolutionarily conserved enzymes that regulate numerous aspects of RNA metabolism, including transcription, splicing, translation and degradation (19). Dysregulation of these critical steps affects the ultimate expression of cellular proteins and consequently determines cell fate (20). With a few exceptions, the biochemical characteristics and functions of the majority of human RNA helicases, including DHX32, have not been well characterized. However, RNA helicase gene dysregulation, including downregulation, upregulation and chromosomal translocation, has been observed in various tumor types (9). DDX1 was reported to be overexpressed in breast cancer and associated with poor survival (21,22). Furthermore, DDX5 was reported to be overexpressed in and to serve an important role in the proliferation of breast cancer cells (23,24). In addition, RNA helicases have been demonstrated to be involved in the interaction and regulation of various cancer-associated molecules (8–12). Therefore, increasing evidence indicates that RNA helicases serve an important role in cancer initiation and progression.
The widespread expression of DHX32, and the high level of similarity between human and murine DHX32, suggests that DHX32 is a functionally important gene (25). DHX32 was observed to be dysregulated in lymphoid malignancies (6). Furthermore, it was observed that DHX32 protein expression is increased in CRC tissue compared with adjacent wild-type tissues (7). DHX32 protein expression was also significantly associated with cancer location, lymph node metastasis, nodal status, differentiation grade and Dukes' stage (7). A recent study by Lin et al (16) indicated that DHX32 promotes the proliferation, migration and invasion of CRC cells, which suggests that abnormal DHX32 expression is associated with tumor development and progression. Meng et al (26) reported that DHX32 interacts with BRCA2 and CDKN1A interacting protein, which is the interacting protein gene of the DNA repair-associated protein breast cancer 2 (BRCA2) and cyclin-dependent kinase inhibitor 1A (CDKN1A). It has been demonstrated that BRCA2 and CDKN1A serve important roles in breast cancer development (27–29), which suggests that DHX32 is associated with breast cancer progression. The results of the present study indicated that the increased expression of DHX32 in breast cancer is significantly associated with an increased clinical stage and histological grade, and a positive lymph node metastasis and Ki-67 expression status, further suggesting that DHX32 serves a critical role in human breast cancer development. Univariate and multivariate analyses revealed that DHX32 may serve as an independent prognostic factor in patients with breast cancer. To the best of our knowledge, this is the first study that has evaluated the association between DHX32 expression and breast cancer prognosis. However, the exact function and underlying mechanism of DHX32-mediated breast cancer progression remains unclear.
In conclusion, the present study demonstrates that DHX32 expression is associated with the development of breast cancer. In addition, increased DHX32 expression was observed to be associated with poor prognosis in breast cancer, suggesting that DHX32 is a novel prognostic marker. However, large-scale studies are required to verify the prognostic value of DHX32 in breast cancer and further research into the tumor-regulatory mechanisms associated with DHX32 is warranted.
Acknowledgements
The present study was supported by the Beijing Natural Science Foundation (grant nos. 7154193 and 7142051), the High Level Technical Talent Development Fund of the Beijing Health System (grant no. 2013-3-052), and the National Key Technology Research and Development Program of the Ministry of Science and Technology of China (grant no. 2013BAI09B03).
References
- 1.Fan L, StrasserWeippl K, Li JJ, St Louis J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM, Goss PE. Breast cancer in China. Lancet Oncol. 2014;15:e279–e289. doi: 10.1016/S1470-2045(13)70567-9. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Kittaneh M, Montero AJ, Glück S. Molecular profiling for breast cancer: A comprehensive review. Biomark Cancer. 2013;5:61–70. doi: 10.4137/BIC.S9455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abdelhaleem M, Maltais L, Wain H. The human DDX and DHX gene families of putative RNA helicases. Genomics. 2003;81:618–622. doi: 10.1016/S0888-7543(03)00049-1. [DOI] [PubMed] [Google Scholar]
- 5.Fuller-Pace FV. DExD/H box RNA helicases: Multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdelhaleem M. The novel helicase homologue DDX32 is down-regulated in acute lymphoblastic leukemia. Leuk Res. 2002;26:945–954. doi: 10.1016/S0145-2126(02)00040-1. [DOI] [PubMed] [Google Scholar]
- 7.Huang C, Liang X, Huang R, Zhang Z. Up-regulation and clinical relevance of novel helicase homologue DHX32 in colorectal cancer. J Exp Clin Cancer Res. 2009;28:11. doi: 10.1186/1756-9966-28-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdelhaleem M. Do human RNA helicases have a role in cancer? Biochim Biophys Acta. 2004;1704:37–46. doi: 10.1016/j.bbcan.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Robert F, Pelletier J. Perturbations of RNA helicases in cancer. Wiley Interdiscip Rev RNA. 2013;4:333–349. doi: 10.1002/wrna.1163. [DOI] [PubMed] [Google Scholar]
- 10.Guil S, Gattoni R, Carrascal M, Abiáán J, Stévenin J, Bach-Elias M. Roles of hnRNP A1, SR proteins, and p68 helicase in c-H-ras alternative splicing regulation. Mol Cell Biol. 2003;23:2927–2941. doi: 10.1128/MCB.23.8.2927-2941.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hönig A, Auboeuf D, Parker MM, O'Malley BW, Berget SM. Regulation of alternative splicing by the ATP-dependent DEAD-box RNA helicase p72. Mol Cell Biol. 2002;22:5698–5707. doi: 10.1128/MCB.22.16.5698-5707.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myöhänen S, Baylin SB. Sequence-specific DNA binding activity of RNA helicase A to the p16INK4a promoter. J Biol Chem. 2001;276:1634–1642. doi: 10.1074/jbc.M004481200. [DOI] [PubMed] [Google Scholar]
- 13.Anderson SF, Schlegel BP, Nakajima T, Wolpin ES, Parvin JD. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat Genet. 1998;19:254–256. doi: 10.1038/930. [DOI] [PubMed] [Google Scholar]
- 14.Schlegel BP, Starita LM, Parvin JD. Overexpression of a protein fragment of RNA helicase A causes inhibition of endogenous BRCA1 function and defects in ploidy and cytokinesis in mammary epithelial cells. Oncogene. 2003;22:983–991. doi: 10.1038/sj.onc.1206195. [DOI] [PubMed] [Google Scholar]
- 15.Alli Z, Ho M, Ackerley C, Abdelhaleem M. Characterization of murine Dhx32. Exp Mol Pathol. 2007;83:115–118. doi: 10.1016/j.yexmp.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 16.Lin H, Liu W, Fang Z, Liang X, Li J, Bai Y, Lin L, You H, Pei Y, Wang F, Zhang ZY. Overexpression of DHX32 contributes to the growth and metastasis of colorectal cancer. Sci Rep. 2015;5:9247. doi: 10.1038/srep09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Torre LA, Bray F, Siegel RL, Ferlay J, LortetTieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 19.Abdelhaleem M. Helicases: An overview. Methods Mol Biol. 2010;587:1–12. doi: 10.1007/978-1-60327-355-8_1. [DOI] [PubMed] [Google Scholar]
- 20.Abdelhaleem M. Over-expression of RNA helicases in cancer. Anticancer Res. 2004;24:3951–3953. [PubMed] [Google Scholar]
- 21.Taunk NK, Goyal S, Wu H, Moran MS, Chen S, Haffty BG. DEAD box 1 (DDX1) expression predicts for local control and overall survival in early stage, node-negative breast cancer. Cancer. 2012;118:888–898. doi: 10.1002/cncr.26352. [DOI] [PubMed] [Google Scholar]
- 22.Germain DR, Graham K, Glubrecht DD, Hugh JC, Mackey JR, Godbout R. DEAD box 1: A novel and independent prognostic marker for early recurrence in breast cancer. Breast Cancer Res Treat. 2011;127:53–63. doi: 10.1007/s10549-010-0943-7. [DOI] [PubMed] [Google Scholar]
- 23.Mazurek A, Luo W, Krasnitz A, Hicks J, Powers RS, Stillman B. DDX5 regulates DNA replication and is required for cell proliferation in a subset of breast cancer cells. Cancer Discov. 2012;2:812–825. doi: 10.1158/2159-8290.CD-12-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D, Huang J, Hu Z. RNA helicase DDX5 regulates microRNA expression and contributes to cytoskeletal reorganization in basal breast cancer cells. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.011932. M111.011932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alli Z, Ackerley C, Chen Y, AlSaud B, Abdelhaleem M. Nuclear and mitochondrial localization of the putative RNA helicase DHX32. Exp Mol Pathol. 2006;81:245–248. doi: 10.1016/j.yexmp.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 26.Meng X, Liu J, Shen Z. Genomic structure of the human BCCIP gene and its expression in cancer. Gene. 2003;302:139–146. doi: 10.1016/S0378-1119(02)01098-3. [DOI] [PubMed] [Google Scholar]
- 27.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 28.Xia W, Chen JS, Zhou X, Sun PR, Lee DF, Liao Y, Zhou BP, Hung MC. Phosphorylation/cytoplasmic localization of p21Cip1/WAF1 is associated with HER2/neu overexpression and provides a novel combination predictor for poor prognosis in breast cancer patients. Clin Cancer Res. 2004;10:3815–3824. doi: 10.1158/1078-0432.CCR-03-0527. [DOI] [PubMed] [Google Scholar]
- 29.Dai M, AlOdaini AA, Arakelian A, Rabbani SA, Ali S, Lebrun JJ. A novel function for p21Cip1 and acetyltransferase p/CAF as critical transcriptional regulators of TGFb-mediated breast cancer cell migration and invasion. Breast Cancer Res. 2012;14:R127. doi: 10.1186/bcr3322. [DOI] [PMC free article] [PubMed] [Google Scholar]