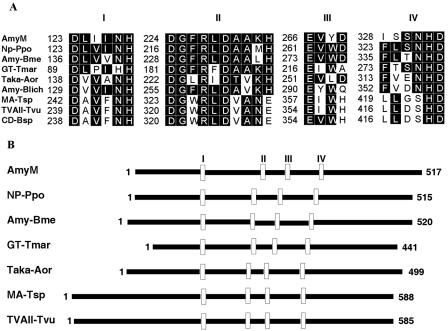
FIG. 2.
Comparisons of amino acid sequences of AmyM with those of related amylases. (A) Amino acid sequences in conserved regions. Highly conserved residues are boxed in black. (B) Schematic alignment of the primary structures. Four conserved regions are marked with open boxes. NP-Ppo, neopullulanase of P. polymyxa (accession no. AAD05199; 47% identity); Amy-Bme, α-amylase of B. megaterium (P20845; 45% identity); GT-Tmar, 4-α-glucanotransferase of Thermotoga maritima (S60618; 33% identity); Taka-Aor, Taka-amylase A of Aspergillus oryzae (P10529; 25% identity); Amy-Blich, α-amylase of Bacillus lichenifomis (P06278; 24% identity); MA-Tsp, maltogenic amylase of Thermus sp. strain IM6501 (AAC15072; 27% identity); TVAII-Tvu, neopullulanase of Thermoactinomyces vulgaris (Q08751; 28% identity); and CD-Bsp, cyclomaltodextrinase of a Bacillus sp. (AAA92925; 27% identity).