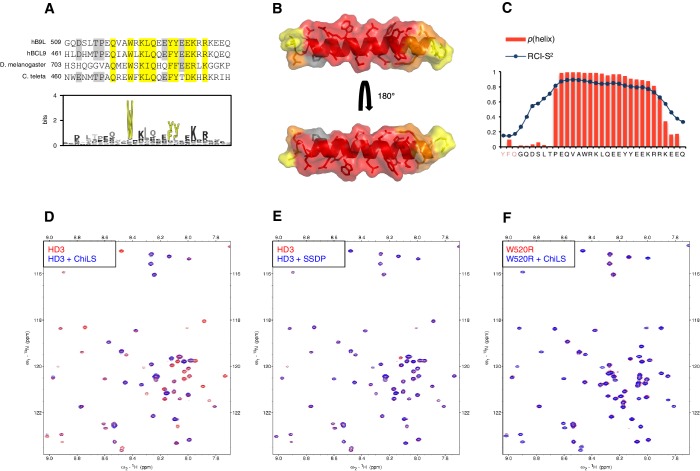
Figure 6. HD3 binds directly to ChiLS.
(A) Top, sequence alignments of HD3; yellow, conserved residues; grey, semi-conserved residues. Bottom, position-specific alignment (HMMER; Finn et al., 2011) for B9L HD3; yellow, conserved tryptophan and phenylalanine/tyrosine doublet. (B) Predicted structure of HD3 (by I-TASSER), with heat-map indicating relative line broadenings upon incubation of 15N-HD3 with ChiLS (see D), ranging from 80% (red) to 40% (yellow); grey, proline (not detectable). (C) TALOS+ predictions of α-helicity of HD3, based on backbone secondary chemical shifts in Figure 6—figure supplement 1; for each position, the rigidity index (RCI-S2) and helical probability (p) are indicated (pink, N-terminal linker residues). (D–F) Overlays of HSQC spectra of 100 μM 15N-labeled (D, E) wt HD3 or (F) W520R mutant alone (red), and probed with 300 μM (D, F) MBP-Chip205-436-Lip-SSDP1-92 or (E) Lip-SSDP1-92 (blue); line broadening owing to ChiLS binding to wt HD3 (D) results in the disappearance of selected resonances in the blue (HD3 + ChiLS) spectrum, revealing the corresponding resonances from the red (HD3-only) spectrum.