Figure 4. Map of pathways potentially involved in FAF1/H. pylori-associated gastric carcinogenesis based on iTRAQ quantification and Western blotting.
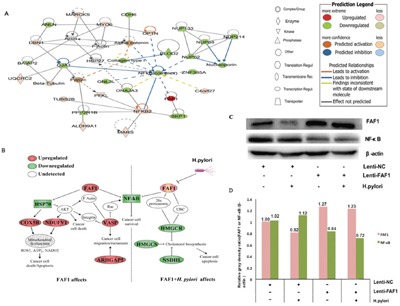
A. Networks of protein inter-relationships were constructed using Ingenuity Pathway Analysis. Red proteins were predicted to be up-regulated, and green proteins to be down-regulated, in FAF1/H. pylori-associated gastric carcinogenesis. B. Protein interaction networks involving FAF1 and H. pylori. Analyzing these networks using the “molecule activity predictor” algorithm in Ingenuity Pathway Analysis generated predictions of the effects of FAF1 up- or down-regulation on downstream molecules. C. Western blotting validation of proteomics results based on different combinations of FAF1 overexpression, NF-κB expression and/or H. pylori infection. D. Image J quantitation of relative FAF1 and NF-κB protein expression in different cell groups. Relative gray density ratios are shown above the bars.
