Figure 4. NF-κB is involved in SRSF3-regulated miR-1908 expression.
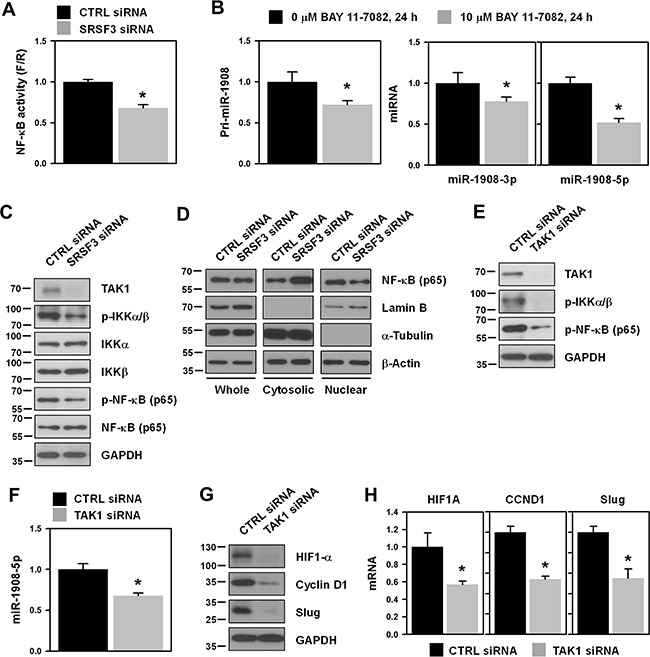
A. U2OS cells were transfected with control (CTRL) or SRSF3-specific siRNA. Transfected cells were resuspended into 12-well plates and then transfected with the NF-κB reporter vector. Transactivation of NF-κB was assessed by calculating the luciferase activity. B. To check whether NF-κB regulates the expression of miR-1908, cells were treated with the NF-κB-specific inhibitor BAY11-7082 (10 μM) for 24 h. The level of miR-1908-3p/5p was determined by RT-qPCR analysis using TaqMan primers. C. Cells were transfected with control (CTRL) or SRSF3-specific siRNA for 48 h and then whole cell lysates were prepared. Western blot analyses were performed using adequate specific antibodies. D. Cells were transfected as described above and then cellular fractionation was performed. Western blot analysis was performed to determine the localization of NF-κB (p65) and α-tubulin and lamin B was checked for cytoplasmic and nuclear markers, respectively. E-F. Cells were transfected with control (CTRL) or TAK1-specific siRNA and then the levels of TAK1 and phosphorylated IKKα/β and NF-κB were assessed by Western blot analysis. The level of miR-1908-5p was determined by RT-qPCR analysis. G-H. The protein and mRNA expression levels of NF-κB target genes including HIF1-α, cyclin D1, and slug, were determined by western blot and RT-qPCR analysis, respectively. All experiments are performed more than three times and data represent the mean ± S.D. Asterisk (*) indicates statistical significance of p<0.05 as determined by Student's t-test.
