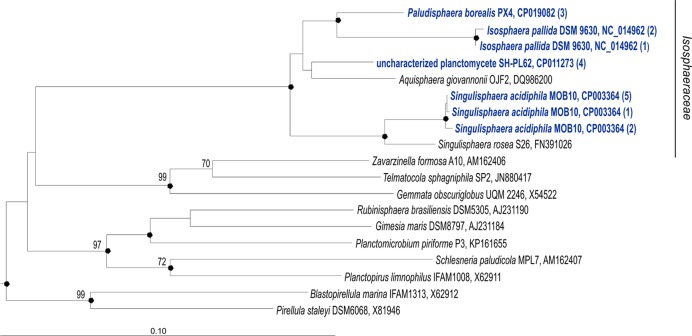
FIGURE 1.
16S rRNA gene-based neighbor-joining tree (Jukes–Cantor correction) showing the phylogenetic position of Paludisphaera borealis PX4T in relation to the three Isosphaeraceae reference organisms used in this study, i.e., Isosphaera pallida IS1BT, Singulisphaera acidiphila DSM 18658T, and undescribed planctomycete strain SH-PL62S (shown in blue). All 16S rRNA gene copies revealed in the genomes of these four planctomycetes are included in the tree. The number of identical 16S rRNA gene copies is shown in parenthesis. Different 16S rRNA gene copies of S. acidiphila DSM 18658T are grouped as follows (locus-tags are given): S. acidiphila (5)—Sinac_3310, Sinac_3707, Sinac_3773, Sinac_5935, Sinac_6238; S. acidiphila (2)—Sinac_2684, Sinac_4695; S. acidiphila (1)—Sinac_5862. Different 16S rRNA gene copies of I. pallida IS1BT are grouped as follows (locus-tags are given): I. pallida (2)— Isop_R0040, Isop_R0051; I. pallida (1)—Isop_R0061. The significance levels of interior branch points obtained in neighbor-joining analysis were determined by bootstrap analysis (1000 data re-samplings). Bootstrap values (1000 data resamplings) of ≥70% are shown. Black circles indicate that the corresponding nodes were also recovered in the maximum-likelihood and maximum-parsimony trees. The root (not shown) was composed of five 16S rRNA gene sequences from anammox planctomycetes (AF375994, AF375995, AY254883, AY257181, and AY254882). Bar, 0.1 substitutions per nucleotide position.