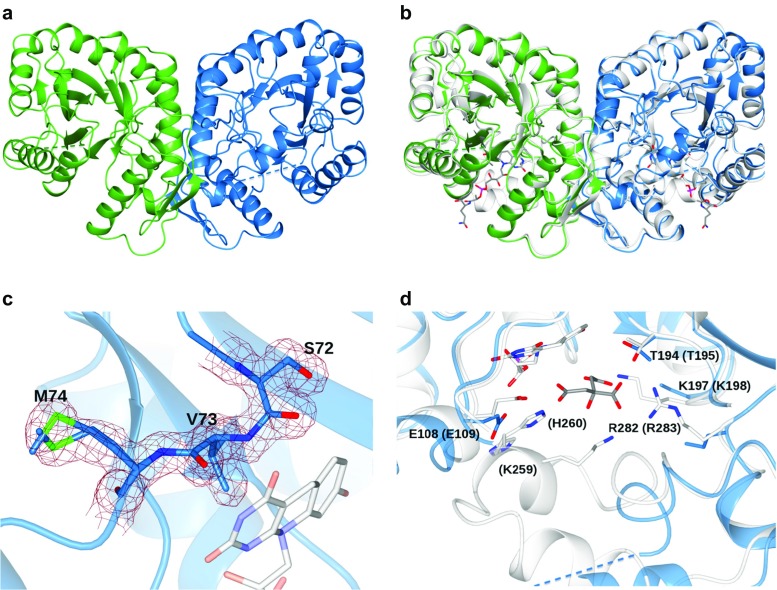
Fig. 5.
Crystal structure of Rh-FGD1 from Rhodococcus jostii RHA1. a Ribbon diagram of the Rh-FGD1 dimer showing the (α/β)8 TIM-barrel architecture of the two monomers colored in light blue (monomer A) and green (monomer B), respectively. The disordered region in each monomer is represented by a dashed line corresponding to residues 254–263 and 250–279 in monomers A and B, respectively. b Superposition of the Rh-FGD1 dimer (colored as in a) onto the homologous Mtb-FGD1 [in white, 84% sequence identity, PDB ID 3Y4B (Bashiri et al. 2008)] with its F420 cofactor bound (carbon, oxygen, nitrogen, and phosphorus atoms in white, red, blue and magenta, respectively). c The nonprolyl cis peptide bond (connecting Ser72 and Val73) and Met74 in a double conformation (sulfur atoms in green) are fitted to the initial 2F o − F c electron density map contoured at 1.2 σ (brown chicken-wire). As a reference, the cofactor F420 from the Mtb-FGD1 structure (superposed as in b) is drawn with shaded colors. d Close-up of the Rh-FGD1 active site superposed to Mtb-FGD1 as in b. The Mtb-FGD1 inhibitor citrate (carbon in gray) is shown bound to the active site. Putative residues involved in substrate binding are labeled with the corresponding Mtb-FGD1 residues in parentheses. The δ, ϵ carbon and ζ nitrogen atoms of K197, and the guanidinium group of R282 side chains were not visible in the electron density and were not included in the final model