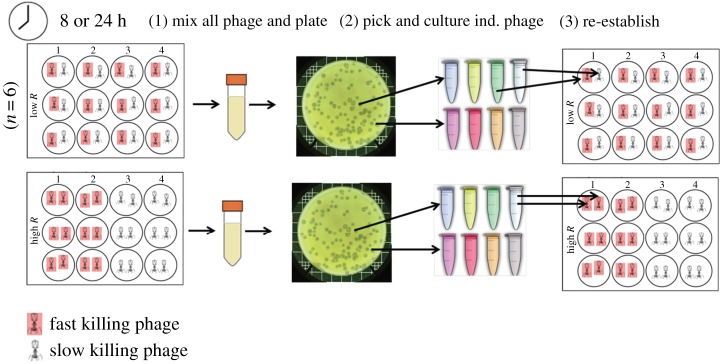
Figure 1.
Experimental set-up. We manipulated viral relatedness and host availability in a fully factorial design. We started the experiment with diverse metapopulations of fast- and slow-killing phage. Half the metapopulations were relatively high relatedness treatments—subpopulations (wells in microtitre plates) founded with single phage clones, while half were relatively low relatedness—founded with two clones. We manipulated host availability by varying the amount of time host-virus populations were cultured: 24 versus 8 h; hosts were a more or less limited resource, meaning that parasite growth was relatively constrained and unconstrained, respectively. (1) At each transfer, we mixed together all subpopulations within a metapopulation and (2) randomly selected virus clones from the plated mixture to (3) re-establish new subpopulations: the output of a subpopulation was consequently dependent on its productivity (global competition).