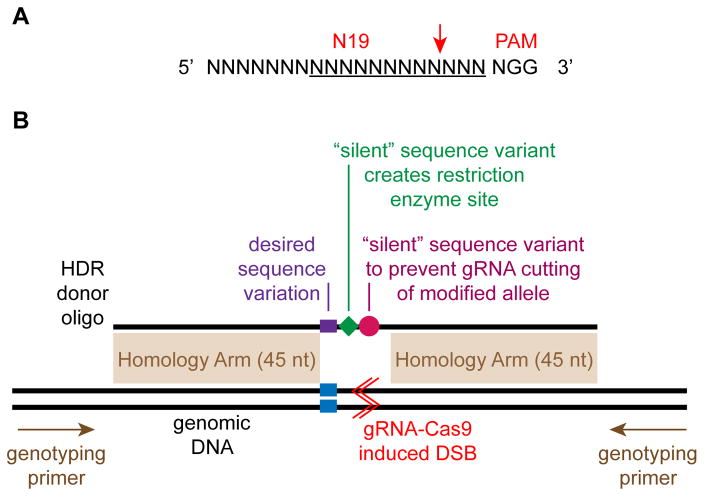
Fig. 5. Design of genome editing reagents.
A. gRNA recognition site. Red arrow indicates the gRNA cleavage site. Underline indicates the 12 nucleotide “seed” sequence that is generally considered most important for Cas9 targeting. The PAM (protospacer adjacent motif) sequence is required for Cas9 targeting. B. A gRNA is designed to introduce a double strand break (DSB, red lines) close to the desired modification site (blue). The HDR donor oligo is design to have homology arms that are at least 45 nt long on either side of the desired sequence modification (purple). Two additional sequence modifications are desirable if they can be achieved without impacting the ultimate experimental goal. First, introduce “silent” sequence variants that eliminate the gRNA recognition sequence after HDR (purple). Second, introduce or remove a restriction enzyme site so that HDR genomes can be identified by restriction digestion of PCR products (green). Genotyping PCR primers are designed flanking the targeted region. These genotyping primers should not overlap the homology arms.