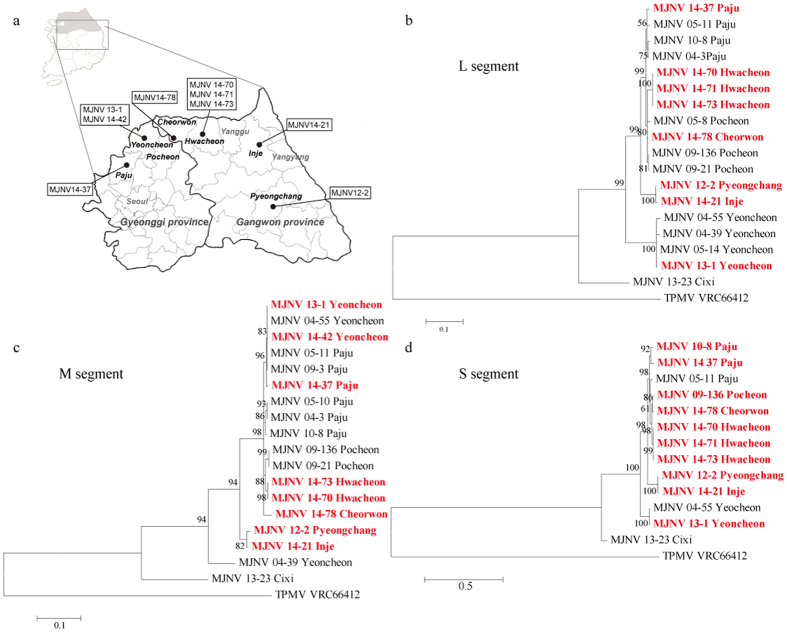
Figure 3. Phylogenetic analyses of the partial sequences of Imjin virus (MJNV) L, M, and S segments in Gangwon and Gyeonggi provinces.
(a) Geographic locations (red circles) of the Republic of Korea indicate where the genomic sequences of MJNV were acquired. Phylogenetic trees were generated by ML methods, based on the (b) 632-nucleotide L segment (coordinates 962–1,593 nt), (c) 531-nucleotide M segment (coordinates 2,252–2,784 nt), and (d) 608-nucleotide S segment (coordinates 405–1,012 nt) of MJNV strains. Red colour indicates the newly obtained MJNV strains during 2011–2014. The phylogenetic positions of MJNV strains are shown in relationship to representative shrew-borne hantaviruses, including Imjin virus from China (MJNV 13–23 Cixi L segment, KJ420567; M segment, KJ420541; S segment, KJ420559) and Thottapalayam virus (TPMV VRC66412 L segment, EU001330; M segment, DQ825771; S segment, NC_010704). (We used Adobe Illustrator CS6 (http://www.adobe.com/products/illustrator.html) to create the map)