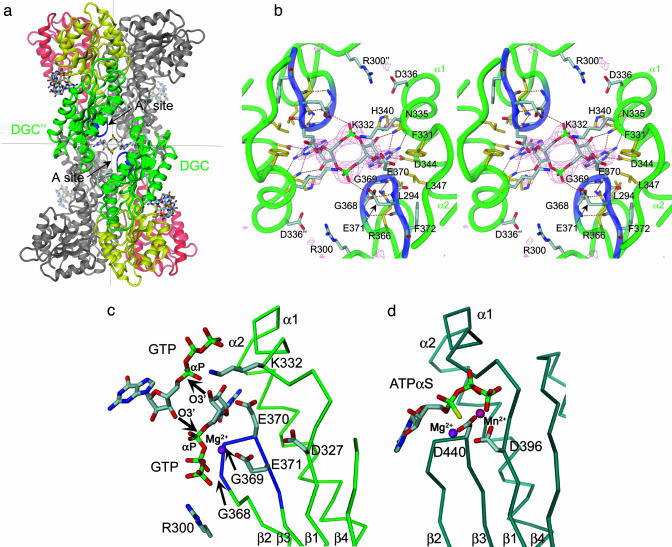
Fig. 2.
Ligand binding to the active site of PleD. (a) Dimers are packed head-to-head to form 222 tetramers. The view is rotated by 135° about the vertical axis with respect to Fig. 1c. The dimers are held together by two c-diGMP ligands that are located on a local two-fold axis of the tetramer (the viewing direction), the ligand molecule in the back has been omitted for clarity. (b) Stereographic close-up view of a. The G368GEEF signature motif comprises residues important for substrate binding (G368, G369, and E371) and catalysis (E370). The omit map of the ligand is contoured at 4 σ. The side chains of E370, E371, R300, K332, and D336 are partly disordered. (c) Complex of DGC with substrate GTP-Mg as modeled on the basis of the product complex shown in b. The positions of the guanine, ribose, and α-phosphoryl moieties are the same as in the product complex structure. The upper substrate, which has been shifted arbitrarily by ≈2 Å to the upper right, would be bound to another two-fold related DGC domain (not shown). The side-chain conformations of E370 and K332 have been adjusted to bring the functional groups into catalytically competent positions. The arrows indicate the nucleophilic attack of the 3′-oxygens on the α-phosphates. (d) AC in complex with substrate analog ATP-α-S [light-blue, PDB ID code 1cjk (25)], view corresponding to that in c. The Cα-trace corresponds to that part of the α-chain, which is structurally homologous to DGC. The β-chain, which provides specific interactions with the adenine base, has been omitted for clarity.