Figure 2.
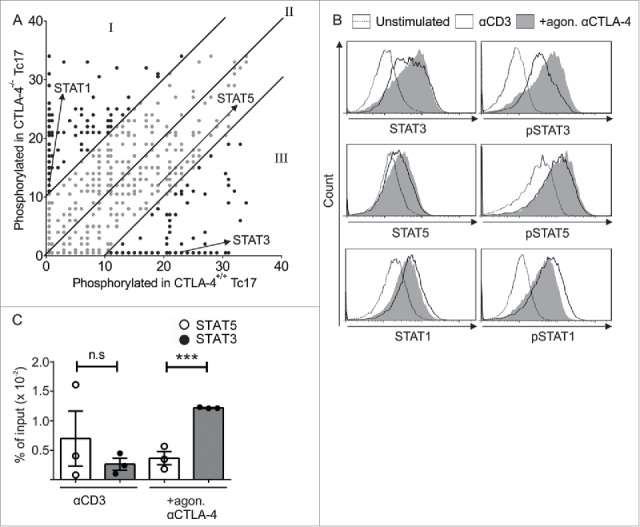
CTLA-4-regulated STAT phosphorylation determines Tc17 differentiation. (A) Dot plot representing the phosphorylation status of the kinase-specific peptide substrates spotted on the PepChip array. Different kinase activities in the lysates from the CTLA-4+/+ and CTLA-4−/− Tc17 cells are shown using a ranking method; each spot represents the extent of phosphorylation of a specific peptide substrate. Using the ranking method, a bisymmetric distribution of peptides is generated, in which phosphorylation was either significantly increased or decreased by CTLA-4 signaling. Peptide substrates that were phosphorylated in the absence of CTLA-4 signaling are reflected by an equivalent peptide with altered or unaltered phosphorylation in response to CTLA-4 signaling. The ranks of the differentially phosphorylated peptides of interest (STAT3, STAT5 and STAT1) are marked by arrows. Spots representing peptides with significantly decreased (I), increased (III) or unaltered (II) phosphorylation as a result of CTLA-4 signaling are shown. (B) Tc17 cells were stimulated with anti-CD3 in the presence or absence of additional CTLA-4 crosslinking for 3 d, harvested, and analyzed for the expression of total and phosphorylated STAT3, STAT1 and STAT5. The data are from a single experiment that is representative of two independent experiments. (C) ChIP analysis of Tc17 cells that were stimulated with CD3 in the presence or absence of additional CTLA-4 crosslinking for 3 d. Tc17 cells were stimulated with IL-6+IL-23 or IL-2 for 30 min, and protein–DNA complexes were crosslinked with formaldehyde and immunoprecipitated with anti-STAT3 or anti-STAT5. The bound DNA was purified and amplified by quantitative PCR with primers designed for the IL-17a promoter site. The results are presented relative to the input DNA. The data are representative of three independent experiments. The error bars denote ± SEM. ***p < 0.001, n.s.: not significant, unpaired t-test.
