Figure 2.
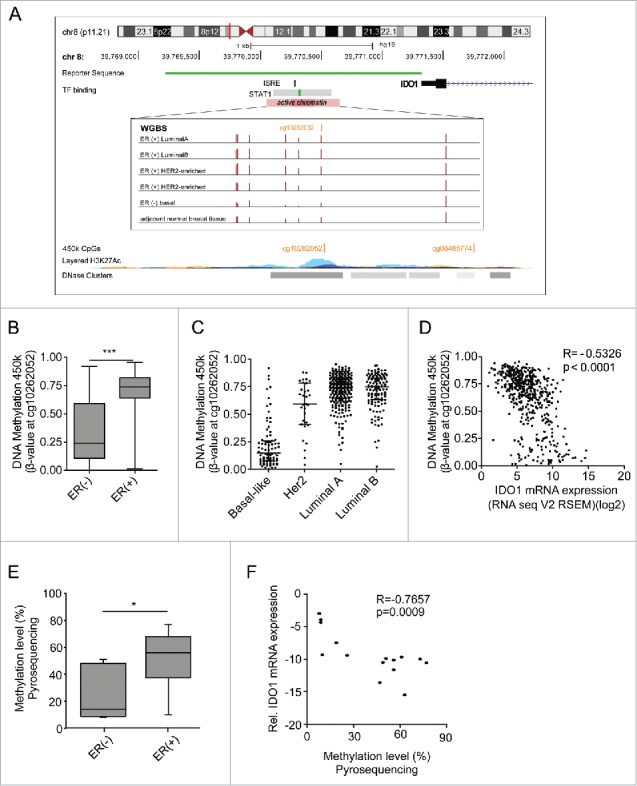
The IDO1 promoter is hypermethylated in ER-positive breast cancer. (A) Schematic representation of regulatory elements in the human IDO1 promoter. In an upstream region of active chromatin (ENCODE ChromHMM E027 and E028, enrichment of H3K27Ac, DNase hypersensitivity cluster) an interferon sensitive response element (ISRE) and a STAT1 binding site overlap with several CpG sites as can be seen in the WGBS data. One of these CpG sites— cg10262052—is covered by 450k arrays (GRCh 37/hg19 assembly). The localization of our IDO1 luciferase reporter gene construct is depicted in green. (B) The DNA methylation level of cg10262052 is significantly lower in ER-negative (n = 99) compared with ER-positive (n = 343) breast cancers (TCGA breast invasive carcinoma, Mann–Whitney U test, ***p < 0.001). Box plots represent the medians and the 75% and 25% percentiles. Whiskers extend to min and max values. (C) DNA methylation of cg10262052 is highest in the ER-positive luminal A, luminal B breast cancer subtypes and lowest in the ER-negative basal-like subtype (basal-like n = 83, Her2-enriched n = 31, luminal A n = 272, luminal B n = 125). (D) The DNA methylation at cg10262052 negatively correlates with IDO1 mRNA expression derived from TCGA breast invasive carcinoma 450k and RNASeq data (n = 511, Spearman's rank correlation). (E) Significant hypermethylation of cg10262052 in ER-positive (n = 9) as compared with ER-negative (n = 6) breast cancer tissue was observed by pyrosequencing (Student's t-test, *p < 0.05). (F) Methylation at this CpG site negatively correlates with IDO1 mRNA expression in these samples (Spearman's rank correlation).
