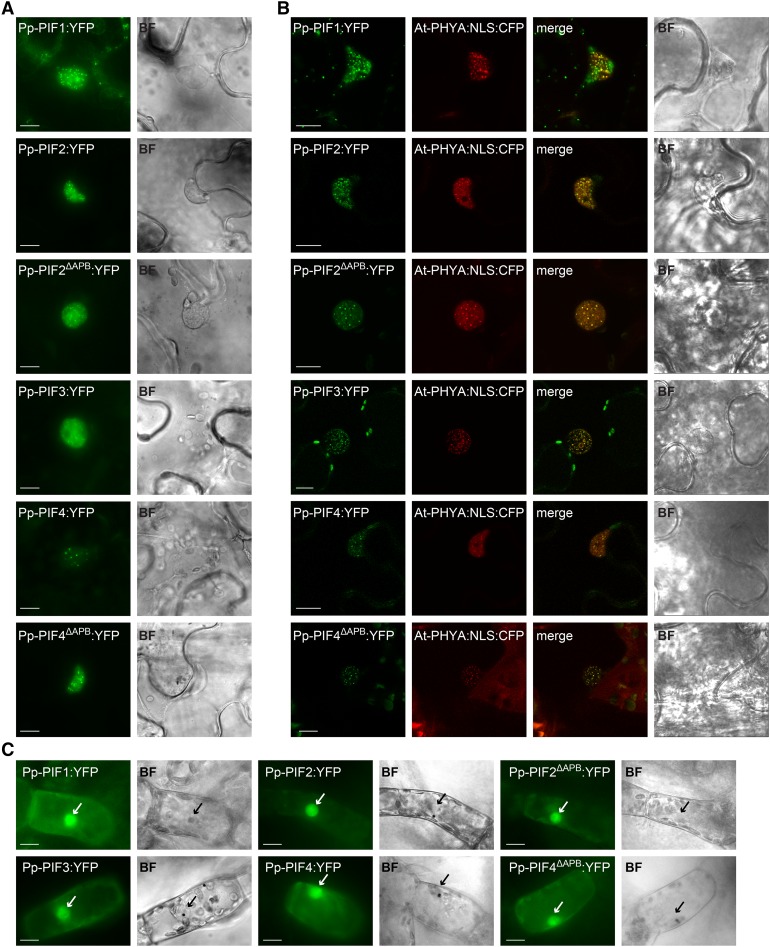
Figure 6.
PIFs from P. patens Localize to the Nucleus.
(A) Pp-PIF1, Pp-PIF2, Pp-PIF2ΔAPB, Pp-PIF3, Pp-PIF4, and Pp-PIF4ΔAPB localize in the nucleus. Leaves of N. benthamiana were transformed by infiltration with Agrobacterium tumefaciens containing a respective Pro35S:Pp-PIF:YFP construct. One day after transformation, the plants were transferred into darkness for another 1 to 3 d before epifluorescence microscopic analysis (n ≥ 20) of epidermal leaf cells. Bar = 10 µm; in all figure parts, BF = bright field.
(B) Pp-PIF1, Pp-PIF2, Pp-PIF2ΔAPB, Pp-PIF3, Pp-PIF4, and Pp-PIF4ΔAPB colocalize with At-PHYA:NLS in the nucleus. Leaves of N. benthamiana were cotransformed with the respective Pro35S:Pp-PIF:YFP and Pro35S:At-PHYA:NLS:CFP. One day after transformation, the plants were transferred into darkness for another 1 to 3 d and analyzed by epifluorescence /confocal microscopy (n ≥ 20; representative confocal images are shown). Bar = 10 µm.
(C) Pp-PIF1, Pp-PIF2, Pp-PIF2ΔAPB, Pp-PIF3, Pp-PIF4, and Pp-PIF4ΔAPB localize to the nucleus in P. patens. Protonema filaments were transiently transformed with the respective Pro35S:Pp-PIF:YFP using particle bombardment and incubated in darkness for 2 to 4 d before epifluorescence microscopy analysis (n ≥ 13). Arrows indicate nuclei. Bar = 10 µm.