Van de Peer and colleagues (Fawcett et al., 2009; Lohaus and Van de Peer, 2016) have written thought-provoking reviews and commentaries on the nonrandom accumulation of ancient polyploidies at the inferred Cretaceous/Tertiary (K/Pg) boundary in the inferred phylogenetic tree of sequenced plants. Their conclusion was that this period of mass extinction—following a meteor hit—created a changed environment that advantaged polyploids and lowered population sizes such that otherwise unlikely survivals took place. They cited recent studies concluding that polyploids may have a unique ability to adapt to stress and offered some traditional population genetics explanations (reviewed in Otto and Whitton, 2000; Conant and Wolfe, 2008; Wendel, 2015) as to why being polyploid increases evolvability. Polyploidy, like sexuality, surely has enhanced plant evolvability some or most of the time.
The Modern Synthesis (Wikipedia has a fine definition) is our traditional “theory of evolution,” marshaling populations and their dynamics, allelic diversity, recombination, and step-by-small-step evolution without a need for new mutation. This traditional logic is called “selectionist” because all needed diversity is presumed to preexist in the population. This letter explores a “mutationist” idea that runs counter to The Modern Synthesis. I propose that extant polyploids have survived as a spandrel, in other words a by-product rather than a direct product, of adaptive selection for asexuality with occasional sex.
WHY ARE POLYPLOID LINEAGES HARD TO KILL?
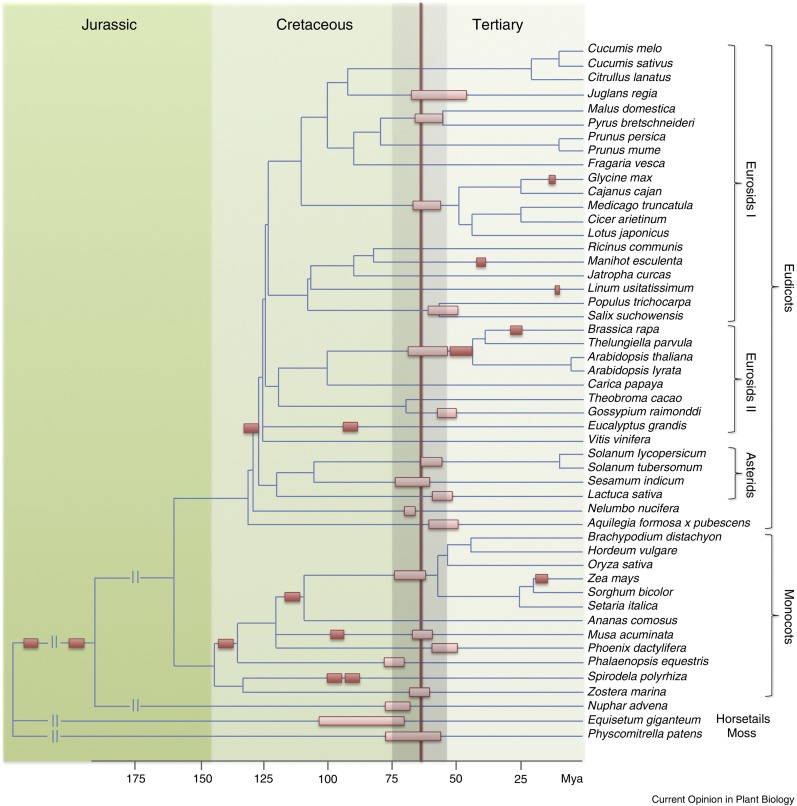
Looking at the Van de Peer group’s evidence from genome sequences (Figure 1), the K/Pg boundary is particularly rich in polyploidy events. Perhaps plant polyploidies occurring at other times are similarly linked to less global or less supported mass extinction events. However, how polyploids might fuel evolvability during bad times is not the only question emerging from Figure 1. There is also the question: Why are some polyploid lineages so hard to kill? Each polyploid lineage surviving to this day can be traced back to approximately one individual polyploidy event happening in one cell from one individual (see section below on the origin of paleopolyploids for a detailed explanation); these are the orange rectangles of Figure 1. Each new polyploid individual has (1) a very low genetic diversity and (2) if reproducing sexually, inaccurate meiosis generating duplication/deficient gametophytes and seeds/kernels (see subsequent section for details.) Comai (2005) presents a thoughtful commentary that highlights these meiotic difficulties. These characteristics should reduce fitness if the organism reproduces sexually. Yet, the polyploid lineages survive while, in almost all cases (Figure 1), related, more genetically diverse lineages go extinct. So, if typical “long-term” survival in plants depends on genetic diversity (Spielman et al., 2004), the long-term survival of paleopolyploidies in plants would be exceptional.
Figure 1.
Schematic Tree of Evolutionary Relationships for Sequenced Plants.
Ancient plant polyploidies (orange rectangles) are distributed with respect to the inferred Cretaceous (K)/Tertiary (Pg) boundary (centered around the vertical brown line). Lighter colored rectangles indicate whole-genome duplications estimated between 55 and 75 million years old (shaded area around the K/Pg boundary). Only lineages that survived to this day are shown. (Reproduced from Lohaus and Van de Peer [2016], http://dx.doi.org/10.1016/j.pbi.2016.01.006, under a Creative Commons Attribution-Noncommercial-No Derivatives License https://creativecommons.org/licenses/by-nc-nd/4.0/.)
Some or all mass extinctions are thought to result from increases in ionizing radiation—from a burst or ozone-depletion—causing mutation, and breakage of DNA or microtubules (Ejzak et al., 2007). A good way for a new polyploid to be hard to kill would be to shield reproductive meristems and avoid meiosis altogether. Plants, unlike most animals, rarely move quickly and need to photosynthesize, so whatever protection evolved must be “in place” under the sun.
Meiosis may or may not play a role in either the origin or maintenance of a polyploid lineage. Vegetative reproduction is widespread among plants and often comes along with perennial phenotype and polyploidy (first edition of Richards [1986]) and Birky [2009]). Comai (2005), in an excellent companion to this letter, suggests that “polyploidy might facilitate the spread of a species by avoiding the need for sexual mates.” Since plants often reproduce vegetatively—perhaps using sex occasionally—a duplicated somatic sector of a budding stem (a runner or a stolon) could take over a bud meristem and generate a polyploid clone of plants, such as a clonal bamboo forest. Duplication results when the spindle checkpoint fails. While vegetative reproduction is near-universal, apomixis (agamospermy; embryos from floral organs) also occurs in flowering plants. As with vegetative reproduction, there is an obvious association of apomixis with polyploidies recent enough to be diagnosed by karyotype (Richards, 1997; Otto and Whitton, 2000; Comai, 2005); all sorts of asexual reproductions are assumed to be similar for this discussion, but see the section below on “Details on the Origin of Paleopolyploidy.” Fractionation, a recombination-based deletion mechanism (Woodhouse et al., 2010), and chromosomal breaks and reunions could happen entirely without sex. Mutations and epimutations could happen without sex; post-sex heterosis fixed in a wide-cross allotetraploid could enhance growth without sex, genome dominance could operate (Freeling et al., 2015a) without sex, and selection operates on all forms of reproduction. Perhaps the reason that polyploids are so hard to kill compared with their nonpolyploid relatives is that the polyploid is protected from the heightened selection pressure of a mass extinction by “hiding out” within the soma of plants that reproduce asexually—especially by budding underground or under water—but have not totally lost the capacity to occasionally develop a flower, fertile or not.
DIPLOIDIZATION OF POLYPLOIDS IS PROMOTED BY “HIDING OUT” IN ASEXUALS
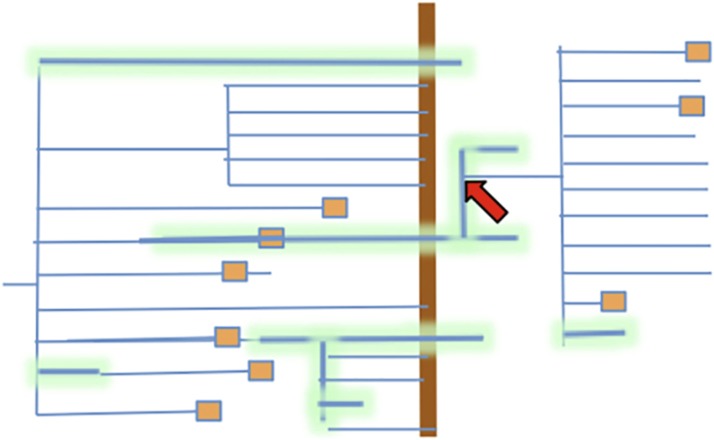
A new polyploid cannot produce competitive, balanced gametophytes until the complicated adaptation called “diploidization” happens (Yant and Bomblies, 2015). Diploidization is the adaptive process of evolving a balanced chromosomal set and alignment machinery that permit efficient segregation and assortment during meiosis. Diploidization is accompanied by chromosome breaks, reunions, and repetitive DNA adjustments; these gross chromosomal changes are probably part of generating “balanced pericentromeres” (Freeling et al., 2015b) necessary for coordinated alignments and movements. We know about some of the mutations that remediate these pairing and movement difficulties. Wild Arabidopsis arenosa is both diploid and a diploidized autotetraploid. Bomblies and colleagues proved that the tetraploid was not preadapted to diploidize and identified most or all of the over 40 loci that must mutate during diploidization (Yant et al., 2013). Comai and colleagues (Henry et al., 2014) identified a single naturally occurring allele in the wild, recent allotetraploid Arabidopsis suecica that increases the meiotic stability of the synthetic (not diploidized) allopolyploid, but diploidization is not complete. Diploidization is best seen as a multistep, mutation-fueled adaptation that might benefit from an environment of very relaxed selection that supports experimentation and absorbs massive failure. A new polyploid reproducing largely somatically (“hiding out”) for “millions of years,” but occasionally flowering—producing many flowers and meiocytes when it does—might occasionally generate a sexual success (red arrow of Figure 2). All data indicate that sexual reproduction has great advantages most of the time. However, in the words of A.K. Richards (Richards, 1986), “It is not even certain that sex alone provides the genetic variation in a population that can fully exploit the diverse niches engendered by any habitat.”
Figure 2.
Hypothetical Tree of Plant Extinction and Survival during a Mass Extinction Event.
An imaginary illustration in the style of Figure 1, but with lineages (blue lines) shown that go extinct as well as the one lineage that is continuous from the nonpolyploid ancestor on the left to the present. If the line is wide with a glow, that lineage reproduces largely asexually. The red arrow marks a rare, successful diploidization event. Note that the thin blue lines, the sexual lineages, do not cross the K/Pg boundary (vertical brown line) and that a nonpolyploid asexual lineage (upper line) does cross the boundary, but then goes extinct.
DETAILS ON THE ORIGIN OF PALEOPOLYPLOIDS
The notion that each polyploid lineage surviving to this day can be traced back to “approximately” one individual (paleo)polyploid event happening in one cell from one individual requires some detailed explanation to be accurate. There are several ways that plants can reproduce asexually and several ways that asexuality and tetraploidy could have co-occurred in the same individual lineage. There is no strong evidence reported in the plant fossil record as to the distribution of asexuals. We are left with inferences based on logic and sometimes based on the assumption that contemporary reproductive behaviors have existed for at least a hundred million years.
All of the paleopolyploidies of Figure 1 can be explained by the most generalized sort of plant asexual reproduction, generically called “vegetative reproduction” (Richards, 1986; chapter 10 of the first but not covered in the second edition). The roots of some plants can generate shoot buds. More generally, the totipotent axillary bud of the repeating module (phytomer) of all plant shoots has the potential to generate new plants via lateral branches, buds off tubers, bulblets, buds off stolons or runners, stem fragments, and similar sources of axillary buds. Those totipotent roots and stems growing underground or underwater offer the reproductive mitotic meristem the most protection against ionizing radiation. (However, underwater meiosis and pollination [Dafni et al., 2012] offer protection as well.) I now use a bamboo—with its stolons, clonal forests, and occasional sex—as my exemplary asexual plant. Almost all perennials and water plants and over half of all extant plant species, reproduce vegetatively, often with occasional sex (Richards, 1986).
If all asexual reproduction were by vegetative reproduction, the data represented in Figure 1 could be explained. Comai (2005) explains the difference between auto- and allopolyploids in extant plants. Recent evidence from genome dominance suggests that ancient plant polyploidies are of both types (Garsmeur et al., 2014). Autotetraploidies would result from diploidizations from somatic organs that had doubled because of a skipped cell division (a failed spindle checkpoint) during mitosis in a diploid floral meristem (with asexuality following) or in a diploid asexual lineage (ramet). Allopolyploids would be similar, but from wide-cross sexual hybrids. A triploid would occur from the sexual hybridization of a diploidized new tetraploid with a sexual diploid, and its duplication could result in a paleohexaploid. There are examples from the paleohexaploid genomes of grape (Vitis vinifera; Lyons et al., 2008) and Brassica rapa (Tang et al., 2012) that support this scenario. Were this simplest way the only way, then all tetraploidies of Figure 1 would reflect one duplication in one cell, and allelic diversity would be zero. However, if multiple asexualization and diploidization events happen at about the same time, or if the plant switches back and forth between these styles, then a single polyploidy could indeed have a population, however limited, at its base.
Some species can reproduce asexually by adventitious embryogenesis from normal diploid or haploid plant floral parts: apomixis (used here as a synonym of agamospermy). Small apomictic populations of diploids and polyploids sometimes form interbreeding complexes called “agamic interbreeding complexes” (Richards, 1997; see Abdi et al. [2016] for a book chapter available online). In such complexes, newly formed polyploids reproduce sexually and asexually (by apomixis), as observed nearly 80 years ago within US species of Crepis (Babcock and Stebbins, 1938). The Panicoid grasses contain a few complexes where diploids are sexual and polyploids are asexual. One of them is called the “diploid-tetraploid-diploid cycle.” Paspalum is one such genus where a breeding population mostly consists of facultative apomictic tetraploids, sexual diploids, triploids, and aneuploids (see Naumova et al. [1999] for a relatively recent reference). Thus, there are populations that could, in theory, help establish a polyploid “event.” That is why the sentence near the beginning of this section reads “approximately one” and not “one” origin for each paleopolyploidy.
Stebbins (1950), commenting on contemporary plant populations, thought that apomixis, like polyploidy, was an evolutionary dead end. While the most abundant types of vegetative reproduction afford maximum protection from ionizing radiation because the reproductive meristem is under ground or underwater, apomixis could be important as well in an environment hostile to chromosomal integrity since meiotic divisions are the most fragile of cell divisions. Even if polyploidy sometimes were to arise from a small population of close relatives—and not in any one individual cell—the allelic diversity of the earliest polyploid of a lineage remains relatively small. That is the point: The successful plant paleopolyploids (Figure 1) dominate the earth using a (near) absence of allelic diversity. Without allelic diversity, current evolutionary theory can falter; traditional population geneticists can but fall back on the Deus ex Machina (god in the machine) of small effective populations. At such desperate times in an evolutionary argument, it’s time to look for a spandrel.
POLYPLOIDY AS A SPANDREL
A “spandrel” used in an evolutionary context is a character acquired automatically because it comes along with a different character that has been selected, but which has no immediate adaptive value in and of itself (Gould and Lewontin, 1979). This “polyploidy is a spandrel of asexuality” hypothesis is illustrated in the imaginary illustration of Figure 2, where lineages are blue lines (as with Figure 1) but, unlike the lines of Figure 1, extinct lineages of the tree are shown as well as the lineage that persist to the present. If reproduction is asexual, the lineage line is wide with a glow; if primarily sexual, the line is thin. Note how all the sexual lineages go extinct (end) near the K/Pg boundary and that the single asexual nonpolyploid lineage crosses as well, but then goes extinct. The arrow marks the diploidization event.
There is nothing continuous about the pattern of ancient polyploidies displayed in Figure 1. Here evolution proceeds via great leaps, with the polyploidy itself being a “systemic mutation” (“a reshuffling or scrambling of the intimate chromosomal architecture” [Goldschmidt, 1953]). These great leaps do not require allelic diversity in the normal sexual sense; what diversity used must have arisen by somatic mutation or epimutation, as with the deletion mutations underlying fractionation, or the chromosomal rearrangements and adaptive mutations that support diploidization. There is no typical population here, no sex, and no obvious way for conventional evolutionary theory to deal with this problem. The mutations enabling diploidization (red arrow of Figure 2), the redevelopment of sexuality, might be called, to use another Goldschmidt term (Goldschmidt, 1953), “macromutations.” The step-by-step mutational fine-tuning associated with breeding regimes, breeding for combinations of micromutations, have nothing to do with solving the problem of the distribution of polyploidies displayed in Figure 1. Nonetheless, evolution of fine-tuning, microevolution, is ever present.
“Diploid” asexuals should also survive mass extinctions. According to Figure 1, several nonpolyploid lineages do cross the K/Pg boundary (e.g., the papaya lineage, an early-diverging Brassicales). However, having survived and been established during stressful times, asexual diploids are expected to be, on average, out-competed during average times by fellow survivors who are polyploid. The idea of an evolutionary spandrel is that the character’s origin is unrelated to the adaptive function this character may exhibit later, once established. The polyploidy reviews cited in this letter all document the many reasons why polyploidy, like sexuality, has been a successful angiosperm character under average or recent environmental conditions.
So, given enough opportunities for mutational trial and error, diploidization might be possible, but rarely, which is consistent with evidence (Arrigo and Barker, 2012). It follows that successful polyploids should rarely occur in existing populations except as a dead end, also consistent with evidence (Stebbins, 1979; Soltis et al., 2014). An unequivocal test of the anti-Stebbins hypothesis that polyploidy accelerates speciation may not be possible with current data (Kellogg, 2016). Schranz et al. (2012) presented some case studies supporting a “lag” between a polyploidy event and an adaptive radiation; that lag would be the asexual period hypothesized here. The lag would be the time it takes for an asexual, new polyploid lineage to generate successful sexual diploids. The observation-based conclusion that wild, extant polyploids might survive because they can reproduce asexually is probably a century old (Wettstein, 1927), but the idea has had little impact in the “why plant polyploidy?” arena.
REVIEW OF ADAPTIVE ADVANTAGES OF ASEXUALITY IN TIMES OF MASS EXTINCTION
An animal might avoid a mass extinction by “hiding out” in a cave. There are genetic, selective consequences of a plant “hiding out” from ionizing radiation in an asexual reproducing underground or underwater. Genes of a polyploid asexual evolve under relatively relaxed selection (1) if they are each one gene of a retained duplicate post-tetraploid pair or (2) to the extent that the gene’s function is confined to sexual reproduction. Genes under relaxed selection (while hiding out) present mutational opportunity to the new, very homozygous asexual polyploid. Some of the mutations the asexual might generate could advance mitotic growth and axillary bud production and could also fuel diploidization. That’s why this spandrel idea is called a “mutationist” evolutionary hypothesis. The permissive “hiding out” period is expected to be a mutational hotbed for the variation necessary to fuel diploidization, the success of the diploid sexual, and the adaptive radiation postdiploidization. If the observations of Schranz et al. (2012), that there may be a lag between a polyploidy and subsequent adaptive radiation, apply to the pre-Cretaceous, this spandrel idea may even help solve Darwin’s “abominable mystery” (Friedman, 2009) as to the source of the burst of floral morphological diversity exhibited by Cretaceous angiosperm fossils.
OBSERVATIONS ON EXTANT PLANTS CAN BE OFF THE POINT
Lest we lose track of the main idea of this letter in the details derived from observing extant plant populations, please consider this: Plant phylogenetic history is not over. If this history repeats, then extant plant lineages—polyploid or not—will go extinct during the next mass extinction with very rare exceptions, these being equivalent to the undiploidized polyploids depicted in Figure 1. According to the idea put forth here, the rare survivors will be (first and foremost) asexual because asexuals are harder to kill, and polyploidy (and perhaps sexual character diversity) will come along only as a spandrel. Once established, the polyploid might now fuel evolution by virtue of its polyploid-specific advantages.
CONCLUSION
Switching the question from “how do polyploids advance evolvability?” to “why are some polyploids so hard to kill?” leads this argument to a spandrel and then to a testable hypothesis. If this “polyploidy as a spandrel of asexuality” hypothesis—an extreme mutationist hypothesis—is correct, our selectionist theory of evolution should be made more inclusive. We know so little about the genetics of asexuality in plants. It is time to learn more about mutations, removal of mutations, epigenetics (for animals, see Castonguay and Angers [2012]), chromatin remodeling, and speciations within plant asexual lineages.
Supplementary Material
Acknowledgments
This work was supported by the NSF Plant Genome Research Program IOS-1546825 to R. Mosher and team. I thank D. Burgess, J. Birchler, R. Mosher, J. Xu, W.Z. Cande, N. Eckardt, and many anonymous reviewers for criticism and suggestions.
AUTHOR CONTRIBUTIONS
M.F. conceived and wrote this letter.
Footnotes
Articles can be viewed without a subscription.
References
- Abdi S., Shashi, Dwivedi A., Bhat V. (2016). Harnessing apomixis for heterosis breeding in crop improvement. In Molecular Breeding of Sustainable Crop Improvement, Ragpal V., Rao S., Raina S., eds (Cham, Switzerland: Springer International Publishing), pp. 79–100. [Google Scholar]
- Arrigo N., Barker M.S. (2012). Rarely successful polyploids and their legacy in plant genomes. Curr. Opin. Plant Biol. 15: 140–146. [DOI] [PubMed] [Google Scholar]
- Babcock E.B., Stebbins G.L. (1938). The American species of Crepis: their relationships and distribution as affected by polyploidy and apomixis. Carnegie Inst. Wash. Publ. 504: 1–200. [Google Scholar]
- Birky C.W.J. (2009). Asexual speciation. In Lost sex: The Evolutionary Biology of Parthanogenesis, Van Dijk P., Martens K., Schon I., eds (Dordrecht, The Netherlands: Springer Science and Business Media; ), pp. 201–216. [Google Scholar]
- Castonguay E., Angers B. (2012). The key role of epigenetics in the persistence of asexual lineages. Genet. Res. Int. 2012: 534–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. (2005). The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6: 836–846. [DOI] [PubMed] [Google Scholar]
- Conant G.C., Wolfe K.H. (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9: 938–950. [DOI] [PubMed] [Google Scholar]
- Dafni A., Hese M., Pacini E. (2012). Pollen and Pollination. (Berlin: Springer Science and Business Media; ). [Google Scholar]
- Ejzak L.M., Melott A.L., Medvedev M.V., Thomas B.C. (2007). Terrestrial consequences of spectral and temporal variability in ionizing photon events. Astrophys. J. 654: 373–384. [Google Scholar]
- Fawcett J.A., Maere S., Van de Peer Y. (2009). Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc. Natl. Acad. Sci. USA 106: 5737–5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M., Scanlon M.J., Fowler J.E. (2015a). Fractionation and subfunctionalization following genome duplications: mechanisms that drive gene content and their consequences. Curr. Opin. Genet. Dev. 35: 110–118. [DOI] [PubMed] [Google Scholar]
- Freeling M., Xu J., Woodhouse M., Lisch D. (2015b). A solution to the C-value paradox and the function of junk DNA: the Genome Balance Hypothesis. Mol. Plant 8: 899–910. [DOI] [PubMed] [Google Scholar]
- Friedman W.E. (2009). The meaning of Darwin’s ‘abominable mystery’. Am. J. Bot. 96: 5–21. [DOI] [PubMed] [Google Scholar]
- Garsmeur O., Schnable J.C., Almeida A., Jourda C., D’Hont A., Freeling M. (2014). Two evolutionarily distinct classes of paleopolyploidy. Mol. Biol. Evol. 31: 448–454. [DOI] [PubMed] [Google Scholar]
- Goldschmidt R.B. (1953). Evolution as viewed by one geneticist. Am. Sci. 40: 84–98. [Google Scholar]
- Gould S.J., Lewontin R.C. (1979). The spandrels of San Marco and the panglossian paradigm: a critique of the adaptionist programme. Proc. R. Soc. Lond. B Biol. Sci. 205: 581–598. [DOI] [PubMed] [Google Scholar]
- Henry I.M., Dilkes B.P., Tyagi A., Gao J., Christensen B., Comai L. (2014). The BOY NAMED SUE quantitative trait locus confers increased meiotic stability to an adapted natural allopolyploid of Arabidopsis. Plant Cell 26: 181–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg E.A. (2016). Has the connection between polyploidy and diversification actually been tested? Curr. Opin. Plant Biol. 30: 25–32. [DOI] [PubMed] [Google Scholar]
- Lohaus R., Van de Peer Y. (2016). Of dups and dinos: evolution at the K/Pg boundary. Curr. Opin. Plant Biol. 30: 62–69. [DOI] [PubMed] [Google Scholar]
- Lyons E., Pederson B., Kane J., Freeling M. (2008). The value of nonmodel genomes and an example using SynMap within CoGe to dissect the paleohexaploidy that preceeds the rosids. Trop. Plant Biol. 1: 181–190. [Google Scholar]
- Naumova T., Hayward M., Wagenvoort M. (1999). Apomixis and sexuality in diploid and tetraploid accessions of Brachiaria decumbens. Sex. Plant Reprod. 12: 43–52. [Google Scholar]
- Otto S.P., Whitton J. (2000). Polyploid incidence and evolution. Annu. Rev. Genet. 34: 401–437. [DOI] [PubMed] [Google Scholar]
- Richards A.J. (1986). Plant Breeding Systems. (London: Allen and Unwin; ). [Google Scholar]
- Richards A.J. (1997). Plant Breeding Systems. (London: Chapman and Hall; ). [Google Scholar]
- Schranz M.E., Mohammadin S., Edger P.P. (2012). Ancient whole genome duplications, novelty and diversification: the WGD Radiation Lag-Time Model. Curr. Opin. Plant Biol. 15: 147–153. [DOI] [PubMed] [Google Scholar]
- Soltis D.E., Visger C.J., Soltis P.S. (2014). The polyploidy revolution then...and now: Stebbins revisited. Am. J. Bot. 101: 1057–1078. [DOI] [PubMed] [Google Scholar]
- Spielman D., Brook B.W., Frankham R. (2004). Most species are not driven to extinction before genetic factors impact them. Proc. Natl. Acad. Sci. USA 101: 15261–15264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins G. (1950). Variation and Evolution in Plants. (New York: Columbia University Press; ). [Google Scholar]
- Stebbins G.L. (1979). Polyploidy in plants: unsolved problems and prospects. Basic Life Sci. 13: 495–520. [DOI] [PubMed] [Google Scholar]
- Tang H., Woodhouse M.R., Cheng F., Schnable J.C., Pedersen B.S., Conant G., Wang X., Freeling M., Pires J.C. (2012). Altered patterns of fractionation and exon deletions in Brassica rapa support a two-step model of paleohexaploidy. Genetics 190: 1563–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J.F. (2015). The wondrous cycles of polyploidy in plants. Am. J. Bot. 102: 1753–1756. [DOI] [PubMed] [Google Scholar]
- Wettstein F. (1927). Die Erscheinung der Heteroploide, besonders im Pflanzenreich. Ergeb. Biol. 2: 311–356. [Google Scholar]
- Woodhouse M.R., Schnable J.C., Pedersen B.S., Lyons E., Lisch D., Subramaniam S., Freeling M. (2010). Following tetraploidy in maize, a short deletion mechanism removed genes preferentially from one of the two homologs. PLoS Biol. 8: e1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L., Bomblies K. (2015). Genome management and mismanagement–cell-level opportunities and challenges of whole-genome duplication. Genes Dev. 29: 2405–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L., Hollister J.D., Wright K.M., Arnold B.J., Higgins J.D., Franklin F.C., Bomblies K. (2013). Meiotic adaptation to genome duplication in Arabidopsis arenosa. Curr. Biol. 23: 2151–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.