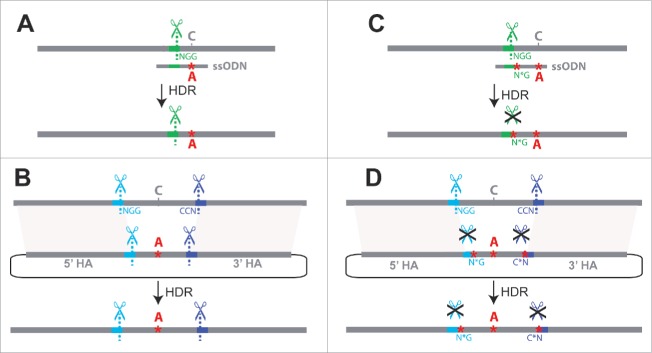
Figure 1.
Challenges for single-stage allele replacement strategies using CRISPR. (A) HDR using a ssODN as repair template is shown. Because ssODNs are limited to ∼200bp, the sgRNA target site (shown in green) must be in close proximity to the sequence to be edited, as the ssODN must span the repair site and have homology to both sides. Unless the introduced change disrupts the target site, the edited locus may be re-cleaved by Cas9, potentially leading to error-prone NHEJ repair. (B) HDR using a dsDNA plasmid as the repair template is shown. If the genomic target site sequences (shown in blue and purple) are present in the donor plasmid, Cas9 cleavage may lead to cutting and subsequent degradation of the donor plasmid; successful repair events will also be vulnerable to cleavage (C) A second mutation at the PAM site (NGG → N*G) may be introduced to prevent re-cleavage of the edited sequence, but this additional mutation(s) may have undesired phenotypic effects. (D) Mutating the PAM sites (NGG → N*G) prevents unwanted cleavage of dsDNA donor or repaired genomic sequence, but involves the addition of extra mutations known as “scars” that may affect phenotypes. Abbreviations used in the figure are defined as follows: NGG = protospacer-adjacent motif; CCN = reverse complement (opposite strand) PAM sequence; N*G or C*N = mutated protospacer-adjacent motif; ssODN = single-stranded oligodeoxynucleotide donor; HDR = Homology-directed repair; HA = Homology arm. Scissor symbols represent target sites expected to be cleaved.