Abstract
In a liver transplant recipient with vancomycin-resistant Enterococcus (VRE) surgical site and bloodstream infection, a combination of pulsed-field gel electrophoresis, multilocus sequence typing, and whole genome sequencing identified that donor and recipient VRE isolates were highly similar when compared to time-matched hospital isolates. Comparison of de novo assembled isolate genomes was highly suggestive of transplant transmission rather than hospital-acquired transmission and also identified subtle internal rearrangements between donor and recipient missed by other genomic approaches. Given the improved resolution, whole-genome assembly of pathogen genomes is likely to become an essential tool for investigation of potential organ transplant transmissions.
Introduction
Donor-derived bacterial infections are a recognized early complication of solid organ transplantation (SOT)[1]. The current definition of transmission requires “clear evidence of the same infection in the donor and at least one of the recipients.”[1] However, common hospital-acquired bacteria such as methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus (VRE) could infect the donor and recipient independently, and it may be difficult to distinguish donor acquisition from hospital acquisition. This is particularly true when infection is not immediately recognized in the recipient. Automated de novo construction of high-quality bacterial genomes using long-read whole genome sequencing (WGS) is a powerful tool that can aid in donor transmission epidemiology[2–5]. In our previous work, WGS approaches were used to confirm donor transmission of MRSA to a liver transplant recipient[6]. Here, we employ multiple genomics strategies, culminating in WGS, to demonstrate donor acquisition of VRE in a liver transplant recipient with surgical site and bloodstream infection by comparing donor and recipient genomes with contemporary hospital isolates.
Case report
Donor
A 51-year-old woman was hospitalized for an exacerbation of systemic lupus erythematosus. The hospital course was complicated by respiratory failure from diffuse alveolar hemorrhage, acute kidney injury requiring renal replacement therapy, and ultimately irreversible central nervous system damage. She was pronounced brain dead and her liver was recovered for transplant. All cultures were negative at this time.
Recipient
A 65-year-old man with hepatitis C cirrhosis and hepatocellular carcinoma was admitted from home and underwent deceased donor liver transplantation. There were no significant intra-operative complications except for expected blood product requirements. The post-operative course was complicated by the development of a complex fluid collection that gradually increased in size; ongoing need for blood transfusion; and acute kidney injury requiring continuous veno-venous hemofiltration (CVVH). Renal function improved and CVVH was discontinued on post-operative day 6. Fever developed immediately after discontinuation of CVVH. Blood cultures obtained on post-operative days 7–9 grew VRE. Linezolid was started on post-operative day 10. The patient was taken to the operating room on post-operative day 13, and one liter of hematoma was evacuated. Cultures of the hematoma grew VRE. Follow-up blood cultures were negative and fever resolved. Linezolid was discontinued thirteen days after evacuation of the hematoma.
Donor blood cultures at the donor hospital eventually grew Gram-positive cocci in pairs and chains and yeast on post-transplant days 1 and 2, respectively. These culture results were finalized on post-transplant day 6 as VRE and Candida glabrata, respectively. The final donor culture results were communicated to our hospital on post-transplant day 15. Caspofungin was started in the recipient at this time and continued for 21 days; however, none of the recipient cultures grew C. glabrata. The recipient was eventually discharged home after a 49-day hospitalization. He has not required any further hospitalizations and is doing well approximately five years later with excellent graft function.
Materials and methods
The donor blood VRE isolate (VRE Donor) was obtained from the donor hospital by LiveOnNY (formerly New York Organ Donor Network) for comparison with the recipient blood isolate (VRE Recipient). Ten additional VRE blood culture isolate strains in the preceding eight weeks from the recipient hospital were retrieved for comparison.
PFGE
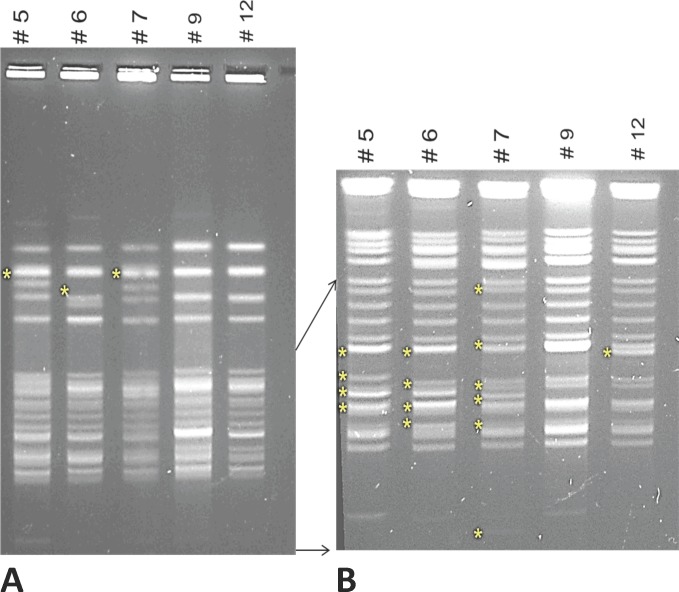
VRE Donor, VRE Recipient and three control (VRE 5, 6, 7) isolates were selected for pulse-field gel electrophoresis (PFGE). Agarose plugs containing genomic DNA were digested with SmaI, and pulsed-field gel electrophoresis (PFGE) was performed using a previously described method[7], but with ramped pulse times of 2 s and 28 s to resolve higher-molecular weight fragments and 2 s and 7s s to resolve low-molecular weight fragments.
PacBio DNA preparation and sequencing
For whole genome sequencing, single bacterial colonies were grown overnight in Luria-Bertani (LB) broth and high molecular weight DNA extraction was performed as previously described[6]. DNA library preparation and Pacific Biosciences (PacBio) sequencing was performed according to the manufacturer’s instructions and reflects the P5-C3 sequencing enzyme and chemistry, respectively. For each isolate, large insert libraries size-selected for fragments greater than 7kb were run on the PacBio RS. A detailed description of the sequencing protocol can be found in S1 File.
Genome assembly, circularizitation, resequencing, annotation
PacBio sequences were assembled using HGAP3 (https://github.com/PacificBiosciences/SMRT-Analysis/wiki/SMRT-Analysis-Software-Installation-v2.2.0). Assemblies were circularized using a custom pipeline employing Nucmer[8]. In short, contig ends that show strong overlap at the boundaries (overlapping alignments exceeding 500 bp) were suggestive of a circular joining site. To eliminate overlapping sequences at the end of contigs and to increase accuracy, the assemblies were joined at the implied overlap point, reoriented (to cause the ends to be internal sequences), and these circularized assemblies were re-sequenced using SMRTpipe 2.2.0. Illumina sequences were mapped to the polished assemblies using bwa-mem[9] (version 0.7.10) in order to verify SNPs and correct assembly errors. Isolates were sequenced to sufficient coverage for de novo assembly on the PacBio RS II. The resulting assemblies were utilized for multilocus sequence typing (MLST) and whole-genome comparative analyses. Table A in S1 File shows sequencing input and assembly quality for all isolates.
Illumina resequencing and variant resolution
In order to further improve assembly quality Illumina resequencing was performed for nine isolates, including VRE Donor and Recipient. Briefly, 0.5–1 μg of input DNA taken from the same aliquot used for Pacbio sequencing was sheared to an average fragment size of 200 bp on a Bioruptor Pico sonicator (Diagenode). Next, amplicon sequence libraries were prepared using the end repair, A-tailing, and adaptor ligation NEBNext DNA library prep modules for Illumina from New England Biolabs according to the manufacturer’s protocol. Following final size-selection with 1x volume Ampure XP beads, and secondary PCR (8 cycles) to introduce barcoded primers, multiplexed libraries were sequenced on the Illumina HiSeq 2500 platform in a single-end 100nt run format.
Sequence errors in the assemblies were corrected by mapping Illumina reads to the assembled contigs with BWA mem[10]. Variants were called using bcftools[11] and then corrected in the assembly. In repetive regions where one repeat contains more errors than the other, BWA mem will map all reads to the copy of the repeat with the least errors. This results in the more erroneous region remaining uncorrected. In order to correct these regions, reads were remapped to sections of the genome with significantly lower than normal coveage (half the median). Indels in these errors were then corrected if a variant was called with bcftools with more than 90% of reads in consensus.
To help resolve potentially missed plasmids, Illumina reads were also assembled using SPAdes[12] version 3.6.0 with default settings. Assembled contigs were then aligned to the PacBio assembly using BLAST+[13]. Illumina contigs that mapped along more than 90% of their length at 90% or greater identity were removed as they are considered to be part of the original PacBio + Illumina assembly. Next, low-coverage non-bacterial contaminant sequences were removed and remaining contigs were circularized using Circlator[14] and annotated by Prokka[15] to identify small plasmids < 10kb (Table B in S1 File). For the donor and recipient genomes complete, circularized assemblies were created by including contigs generated from both the illumina and PacBio assemblies. NCBI accession numbers for deposited assemblies are contained in Table C in S1 File.
MLST and vancoymcin typing
MLST typing was performed by uploading whole genome assemblies to http://pubmlst.org, a public database of MLST types, which includes data for Enterococcus faecium and by using core genome MLST (cgMLST) as part of Ridom SeqSphere+ (http://www.ridom.com/seqsphere/cgmlst/). The pubMLST database returned an MLST type for each genome and types for each allele in seven genes: adk, atpA, ddl, gdh, gyd, pstS and purK; these types were confirmed by SeqSphere for all strains except VRE1, for which no MLST type was given (Table D in S1 File). A subset (10) genomes were further verified using PCR and Sanger sequencing with 9/10 confirming the predicted MLST type obtained from WGS. The only differing strain type was VRE11; Sanger sequencing of VRE11 differed in the atpA and ddl gene assignments which yielded an MLST type that has not previously been observed (potentially an error in the Sanger result).
To determine the vancomycin resistance genotype, PacBio amino acid sequences were analyzed with CARD’s RGI software (3.0.9) using default settings[16]. Initial results indicated an incomplete van operon in VRE 11 missing vanHA and vanXA, respectively. Results were therefore cross-referenced with Illumina nucleotide sequences which identified a complete vancomycin resistance operon. Initial results also indicated vanB ligase genes occurring in some genomes. All CARD reported vancomycin genes were run through BLAST to confirm vanA ligase identity. All isolates were tested with Vitek; genotypes and MIC values are provided in Table D in S1 File.
Comparison between strains and phylogenetic analysis
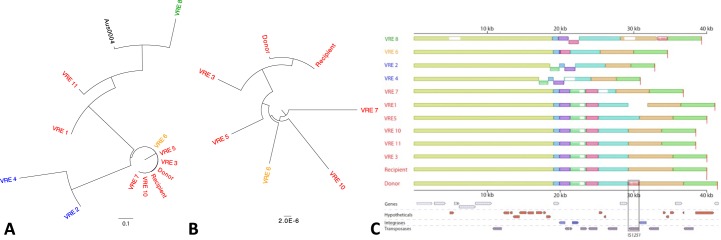
Phylogenetic analyses was performed using HarvestTools v1.1.2[17]. First Parsnp v1.2 was used to obtained core genomes from our Pacbio assemblies, utilizing the Aus0004 genome as reference[18] and filtering for PhiPack identified regions of recombination[19]. Prokka[20] was used for genome annotations and Roary v3.6.1[21] was used to calculate core gene set sizes. De novo assemblies were also to examined for potential structural differences via Nucmer alignment (MUMmer version 3.23)[8]. Lastly, Progressive Mauve[22] was used to create the multiple sequence alignments shown in Fig 1B.
Fig 1. Genomic comparison of VRE isolates.
(A) Conserved sequence blocks generated by HarvestTools 1.1.2 to construct the phylogenetic tree. (B) A second phylogeny was performed on more closely related strains, to refine the Recipient-Donor clade. Isolates with the hypervariable groupings shown in (C) share the same color in both phylogenies. (C) A 40kb interval from the donor strain spanning the hypervariable chromosomal locus was extracted and homologous sequence blocks were obtained from each isolate. Each colored block corresponds to syntenic interval between strains, and isolates are grouped by primary syntenic block order. The red recipient block (outlined in black) corresponds to a mobile element gene insertion of an IS1251-like element. The corresponding genes in the Recipient genomic interval are shown below.
Results
PFGE
VRE Donor and VRE Recipient had almost identical SmaI restriction enzyme PFGE patterns, differing only in the presence of a single brighter band (Fig 2). The control strains were considerably different, both from each other and from VRE Donor and VRE Recipient, showing greater than 6 band changes (Fig 2) following Tenover et al. criteria[23].
Fig 2. Agarose gel showing SmaI digestion patterns of strains via PFGE.
(A) PFGE gel of VRE 5, 6, 7, VRE Recipient and VRE Donor isolates with ramped pulse times of 2s to 28s to resolve higher-molecular weight fragments. (B) Same as A but with ramped pulse times of 2s to 7s to resolve low-molecular weight fragments. Asterisks (*) indicate missing or variable DNA band sizes among isolates. Arrows indicate the corresponding higher resolution DNA band area in panel B relative to panel A.
MLST
While VRE Donor and Recipient had the same ST736 MLST genotype, this genotype was shared with three other control isolates from the recipient hospital: VRE 5, VRE 6, and VRE 7. Nearly all remaining isolates represented distinct MLSTs with genotypes that have been identified worldwide and seem to be widely distributed[24–29].
Whole genome comparison
The de novo assembled VRE genomes were highly contiguous; in all cases, the assemblies contained fewer than 10 contigs with the largest contig representing more than 50% of the total genome length. The isolates were quite diverse, containing a core genome of 2161 genes with an average of 3140 genes per genome. Fig 1A shows a phylogenetic tree representing SNP distances between the 12 VRE strains based on whole genome alignment (Methods). The resulting phylogeny identified a Donor-Recipient specific subclade within the ST736 MLST subtype. Pairwise whole genome comparison was then performed between VRE Donor and VRE Recipient which were assembled to completion. While the two isolates were nearly identical at the SNV level, containing a single SNV within the main chromosome, they were not completely structurally identical: Four intervals exhibited inserted or deleted sequence between the isolates (Figure A and Table E in S1 File), and a 1.5kb plasmid interval contained 15 intergenic SNVs. All of the inserted or deleted sequences contained tranposases with similarity to known insertion sequences (ISs), IS1251[30] and ISEfa11[31], previously associated with the vanA gene cluster. The insertion with high similarity to IS1251 was missing from two locations (one chromosomal and one plasmid) within the VRE Donor. The corresponding chromosomal location was interrogated across all 12 isolates by isolating a 40kb flanking interval that encodes 29 genes including 10 mobile elements. Fig 1B and Table F in S1 File highlight the mobile insertion between donor and recipient. Notably, the orthologous region was structurally variable across nearly all isolates. Even those genomes with similar organization and block size, such as VRE 10 and VRE 11, contained multiple SNVs between one another. Based on ordering and directionality of syntenic blocks, these strains were separated into four categories (colors of isolates in Fig 1A); however, local variation (such as insertions and deletions and SNVs) still existed within each group. Closest strain pairs on the SNV phylogeny did not necessarily correspond to the closest structural matches for this region (e.g. VRE10 and VRE11), and in some cases large-scale rearrangements had occurred between two proximal isolates (e.g. VRE11 and VRE8). Together, this suggests that this region is likely to mutate or undergo rearrangement over a short period (as in the case of VRE Donor and VRE Recipient).
Discussion
VRE surgical site and bloodstream infection in a hospitalized liver transplant recipient meets the definition of a hospital-acquired infection[32], but early bacterial infection may also be donor-transmitted. In our case, the possibility of donor-transmitted infection was only considered after the donor infection history was eventually obtained. By genomic analysis of VRE blood isolates from the donor, recipient, and control isolates from the recipient hospital, we were able to demonstrate that this is a likely case of donor transmission rather than hospital acquisition.
Our combination of short and long read genomic approaches highlights the increased specificity of WGS for resolving both SNV and structural differences between similar isolates. All genomic analyses revealed a strong donor and recipient grouping; however, multiple isolates had MLST types consistent with the donor and recipient. And, while PFGE did show a tighter grouping between donor and recipient, only the full de novo assembly was able to clarify the unique structural differences between the donor and recipient isolate. Interestingly, this and related insertion elements have previously been associated with increased antibiotic resistance in the context of the vanA gene cluster[33]. This type of transposase and mobile element variation is consistent with previous studies which have suggested that recombinational exchange is five times more likely to generate new alleles in E. faecium[34]. The high-rates of recombination seen in E. faecium have been shown in simulation to render MLST phylogenetic inference inaccurate[35]. In fact, large structural changes could also cause substantial band shifts or amplitude changes in PFGE profiles. Our data suggest that WGS may be increasingly necessary to unambiguously confirm transmission for structurally mutable genomes. Previous transmission studies define allowed numbers of mutations between two patients over a given time window based on molecular clocks[36], but such analyses are typically restricted to SNVs. In part, this is due to the ubiquity of short-read sequencing which are ideal for reference-based mapping but do not easily facilitate discovery of large scale structural variants insertions and rearrangements. As long-read sequencing becomes more readily available, and automated assembly algorithms continue to improve, it will become increasingly necessary to fold-in large-scale structural changes into genomic models of transmission.
In summary, WGS provided substantial evidence that VRE infection in our liver transplant recipient was not hospital acquired as would have typically been considered but rather transmitted from the deceased donor. We expect that WGS and assembly of pathogen genomes will be increasingly important not only for understanding pathogen biology and evolution but also become a routine and essential tool for investigation of potential organ transplant transmissions in many settings. Our case also highlights the importance of communicating deceased donor culture information to transplant recipient centers so that potential preventive or preemptive strategies can be implemented as early as possible.
Supporting information
(DOCX)
Acknowledgments
We thank Simon Daefler, MD for performing confirmatory MLST and helpful discussions. Additionally, analyses were supported in part through the computational resources and staff expertise provided by the Department of Scientific Computing at the Icahn School of Medicine at Mount Sinai.
Abbreviations
- VRE
vancomycin-resistant Enterococcus
- WGS
whole genome sequencing
- MLST
multilocus sequence typing
- PFGE
pulsed-field gel electrophoresis
- SOT
solid organ transplantation
- MRSA
methicillin-resistant Staphylococcus aureus
- CVVH
continuous veno-venous hemofiltration
- SNV
single nucleotide variation
Data Availability
All data are available from the NCBI with the accession numbers: VRE 1 Assembly CP012430-CP012435 VRE 2 Assembly CP012436-CP012439 VRE 3 Assembly CP012440-CP012446 VRE 4 Assembly CP012447-CP012453 VRE 5 Assembly CP012454-CP012459 VRE 6 Assembly LIVR00000000 VRE 7 Assembly CP012460-CP012464 VRE 8 Assembly CP012465-CP012470 Recipient Assembly CP018825-CP018829 VRE 10 Assembly CP012471-CP012474 VRE 11 Assembly LIHD00000000 Donor Assembly CP018830-CP018834 VRE 2 Illumina Reads SRR3115443 VRE 2 Illumina Reads SRX1543354 VRE 3 Illumina Reads SRR3115446 VRE 3 Illumina Reads SRX1543363 VRE 4 Illumina Reads SRR3115450 VRE 4 Illumina Reads SRX1543364 VRE 5 Illumina Reads SRR3115453 VRE 5 Illumina Reads SRX1543365 VRE 7 Illumina Reads SRR3115512 VRE 7 Illumina Reads SRX1543420 VRE 8 Illumina Reads SRR3115534 VRE 8 Illumina Reads SRX1543443 Recipient Illumina Reads SRR3115540 Recipient Illumina Reads SRX1543444 VRE 10 Illumina Reads SRR3115312 VRE 10 Illumina Reads SRX1543246 Donor Illumina Reads SRR3115319 Donor Illumina Reads SRX1543251 VRE 1 PacBio Reads SRX1258250 VRE 2 PacBio Reads SRX1541965 VRE 3 PacBio Reads SRX1542020 VRE 4 PacBio Reads SRX1542021 VRE 5 PacBio Reads SRX1542071 VRE 6 PacBio Reads SRX1542072 VRE 7 PacBio Reads SRX1542107 VRE 8 PacBio Reads SRX1542146 Recipient PacBio Reads SRX1542110 VRE 10 PacBio Reads SRX1542146 VRE 11 PacBio Reads SRX1542424 Donor PacBio Reads SRX1542425
Funding Statement
This work was supported by the Icahn Institute for Genomics and Multiscale Biology, and also in part by the National Institute of Allergy and Infectious Diseases, grant R01AI119145 and NRSA Institutional Research Training Grant (5 T32 AI 7647-13) for Global Health Research (DRA), and F30AI122673. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ison MG, Grossi P. Donor-derived infections in solid organ transplantation. Am J Transplant. 2013;13 Suppl 4: 22–30. Available: http://www.ncbi.nlm.nih.gov/pubmed/23464995 [DOI] [PubMed] [Google Scholar]
- 2.Bashir A, Klammer AA, Robins WP, Chin C-S, Webster D, Paxinos E, et al. A hybrid approach for the automated finishing of bacterial genomes. Nat Biotechnol. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved.; 2012;30: 701–7. 10.1038/nbt.2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, Ganapathy G, et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved.; 2012;30: 693–700. 10.1038/nbt.2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved.; 2013;10: 563–9. 10.1038/nmeth.2474 [DOI] [PubMed] [Google Scholar]
- 5.Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med. 2011;365: 709–17. 10.1056/NEJMoa1106920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altman DR, Sebra R, Hand J, Attie O, Deikus G, Carpini KWD, et al. Transmission of Methicillin-Resistant Staphylococcus aureus via Deceased Donor Liver Transplantation Confirmed by Whole Genome Sequencing. Am J Transplant. 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murray BE, Singh K V., Heath JD, Sharma BR, Weinstock GM. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28: 2059–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delcher AL, Salzberg SL, Phillippy AM. Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics. 2003;Chapter 10: Unit 10.3. [DOI] [PubMed]
- 9.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009/05/20. 2009;25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013; Available: http://arxiv.org/abs/1303.3997
- 11.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27: 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. Mary Ann Liebert, Inc.; 2012;19: 455–77. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10: 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunt M, Silva N De, Otto TD, Parkhill J, Keane JA, Harris SR. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol. 2015;16: 294 10.1186/s13059-015-0849-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30: 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 16.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57: 3348–3357. 10.1128/AAC.00419-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15: 524 10.1186/s13059-014-0524-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lam MMC, Seemann T, Bulach DM, Gladman SL, Chen H, Haring V, et al. Comparative analysis of the first complete Enterococcus faecium genome. J Bacteriol. 2012;194: 2334–2341. 10.1128/JB.00259-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172: 2665–2681. 10.1534/genetics.105.048975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30: 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 21.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. Oxford University Press; 2015;31: 3691–3. 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14: 1394–1403. 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tenover FC, Arbeit RD, Goering R V, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33: 2233–9. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=228385&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palazzo IC V, Pitondo-Silva A, Levy CE, da Costa Darini AL. Changes in vancomycin-resistant Enterococcus faecium causing outbreaks in Brazil. J Hosp Infect. 2011;79: 70–74. 10.1016/j.jhin.2011.04.016 [DOI] [PubMed] [Google Scholar]
- 25.Ochoa SA, Escalona G, Cruz-Córdova A, Dávila LB, Saldaña Z, Cázares-Domímguez V, et al. Molecular analysis and distribution of multidrug-resistant Enterococcus faecium isolates belonging to clonal complex 17 in a tertiary care center in Mexico City. BMC Microbiol. 2013;13: 291 10.1186/1471-2180-13-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCracken M, Wong A, Mitchell R, Gravel D, Conly J, Embil J, et al. Molecular epidemiology of vancomycin-resistant enterococcal bacteraemia: Results from the canadian nosocomial infection surveillance program, 1999–2009. J Antimicrob Chemother. 2013;68: 1505–1509. 10.1093/jac/dkt054 [DOI] [PubMed] [Google Scholar]
- 27.Damani A, Klapsa D, Panopoulou M, Spiliopoulou I, Pantelidi K, Malli E, et al. A newly described vancomycin-resistant ST412 Enterococcus faecium predominant in Greek hospitals. Eur J Clin Microbiol Infect Dis. 2010;29: 329–331. 10.1007/s10096-009-0847-9 [DOI] [PubMed] [Google Scholar]
- 28.da Silva LPP, Pitondo-Silva A, Martinez R, da Costa Darini AL. Genetic features and molecular epidemiology of Enterococcus faecium isolated in two university hospitals in Brazil. Diagn Microbiol Infect Dis. 2012;74: 267–71. Available: http://www.sciencedirect.com/science/article/pii/S0732889312003288 10.1016/j.diagmicrobio.2012.07.012 [DOI] [PubMed] [Google Scholar]
- 29.Freitas AR, Novais C, Tedim AP, Francia MV, Baquero F, Peixe L, et al. Microevolutionary events involving narrow host plasmids influences local fixation of vancomycin-resistance in Enterococcus populations. PLoS One. 2013;8: e60589 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3612099&tool=pmcentrez&rendertype=abstract 10.1371/journal.pone.0060589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Handwerger S, Skoble J, Discotto LF, Pucci MJ. Heterogeneity of the vanA gene cluster in clinical isolates of enterococci from the northeastern United States. Antimicrob Agents Chemother. 1995;39: 362–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.López M, Sáenz Y, Alvarez-Martínez MJ, Marco F, Robredo B, Rojo-Bezares B, et al. Tn1546 structures and multilocus sequence typing of vanA-containing enterococci of animal, human and food origin. J Antimicrob Chemother. Oxford University Press; 2010;65: 1570–5. 10.1093/jac/dkq192 [DOI] [PubMed] [Google Scholar]
- 32.Garner JS, Jarvis WR, Emori TG, Horan TC, Hughes JM. CDC definitions for nosocomial infections, 1988. Am J Infect Control. 1988;16: 128–140. [DOI] [PubMed] [Google Scholar]
- 33.Handwerger S, Skoble J. Identification of chromosomal mobile element conferring high-level vancomycin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1995;39: 2446–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Homan WL, Tribe D, Poznanski S, Li M, Hogg G, Spalburg E, et al. Multilocus sequence typing scheme for Enterococcus faecium. J Clin Microbiol. 2002;40: 1963–71. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=130786&tool=pmcentrez&rendertype=abstract 10.1128/JCM.40.6.1963-1971.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner KME, Hanage WP, Fraser C, Connor TR, Spratt BG. Assessing the reliability of eBURST using simulated populations with known ancestry. BMC Microbiol. 2007;7: 30 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1865383&tool=pmcentrez&rendertype=abstract 10.1186/1471-2180-7-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Didelot X, Eyre DW, Cule M, Ip CLC, Ansari MA, Griffiths D, et al. Microevolutionary analysis of Clostridium difficile genomes to investigate transmission. Genome Biol. 2012;13: R118 10.1186/gb-2012-13-12-r118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All data are available from the NCBI with the accession numbers: VRE 1 Assembly CP012430-CP012435 VRE 2 Assembly CP012436-CP012439 VRE 3 Assembly CP012440-CP012446 VRE 4 Assembly CP012447-CP012453 VRE 5 Assembly CP012454-CP012459 VRE 6 Assembly LIVR00000000 VRE 7 Assembly CP012460-CP012464 VRE 8 Assembly CP012465-CP012470 Recipient Assembly CP018825-CP018829 VRE 10 Assembly CP012471-CP012474 VRE 11 Assembly LIHD00000000 Donor Assembly CP018830-CP018834 VRE 2 Illumina Reads SRR3115443 VRE 2 Illumina Reads SRX1543354 VRE 3 Illumina Reads SRR3115446 VRE 3 Illumina Reads SRX1543363 VRE 4 Illumina Reads SRR3115450 VRE 4 Illumina Reads SRX1543364 VRE 5 Illumina Reads SRR3115453 VRE 5 Illumina Reads SRX1543365 VRE 7 Illumina Reads SRR3115512 VRE 7 Illumina Reads SRX1543420 VRE 8 Illumina Reads SRR3115534 VRE 8 Illumina Reads SRX1543443 Recipient Illumina Reads SRR3115540 Recipient Illumina Reads SRX1543444 VRE 10 Illumina Reads SRR3115312 VRE 10 Illumina Reads SRX1543246 Donor Illumina Reads SRR3115319 Donor Illumina Reads SRX1543251 VRE 1 PacBio Reads SRX1258250 VRE 2 PacBio Reads SRX1541965 VRE 3 PacBio Reads SRX1542020 VRE 4 PacBio Reads SRX1542021 VRE 5 PacBio Reads SRX1542071 VRE 6 PacBio Reads SRX1542072 VRE 7 PacBio Reads SRX1542107 VRE 8 PacBio Reads SRX1542146 Recipient PacBio Reads SRX1542110 VRE 10 PacBio Reads SRX1542146 VRE 11 PacBio Reads SRX1542424 Donor PacBio Reads SRX1542425