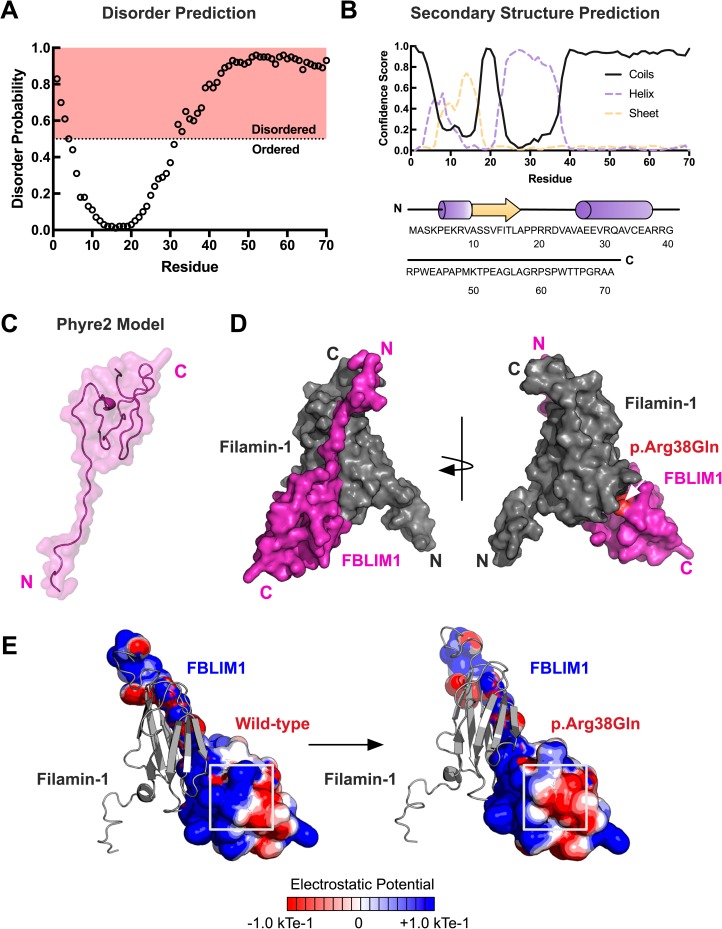
Fig 4. Structural modeling of patient mutation in FBLIM1.
(A) Disorder prediction of the Filamin-1 binding region of FBLP1 (residues 1–70) was performed with DISOPRED version 3. Disorder probability scores greater than 0.5 are considered to be disordered. The R38Q mutation falls in a predicted disordered region. (B) Secondary structure prediction was performed using PSI-PRED. The confidence scores for coils, helices, and strands are shown. (C) The output model of the Filamin-1 binding region of FBLIM1 generated in Phyre2. A total of 86% of the residues are predicted to be disordered, resulting in 76% of the residues having a low confidence score (<90%). Residues 1–24 are predicted to be ordered. (D) Superimposition of the Phyre2 model onto the NMR structure of the Filamin-1-FBLIM1 complex. Modeling of the complex places the R38Q mutation downstream of the interaction site. (E) Electrostatic potential calculations in APBS reveal a loss of positive charge in FBLP1 as a result of the CRMO mutation.