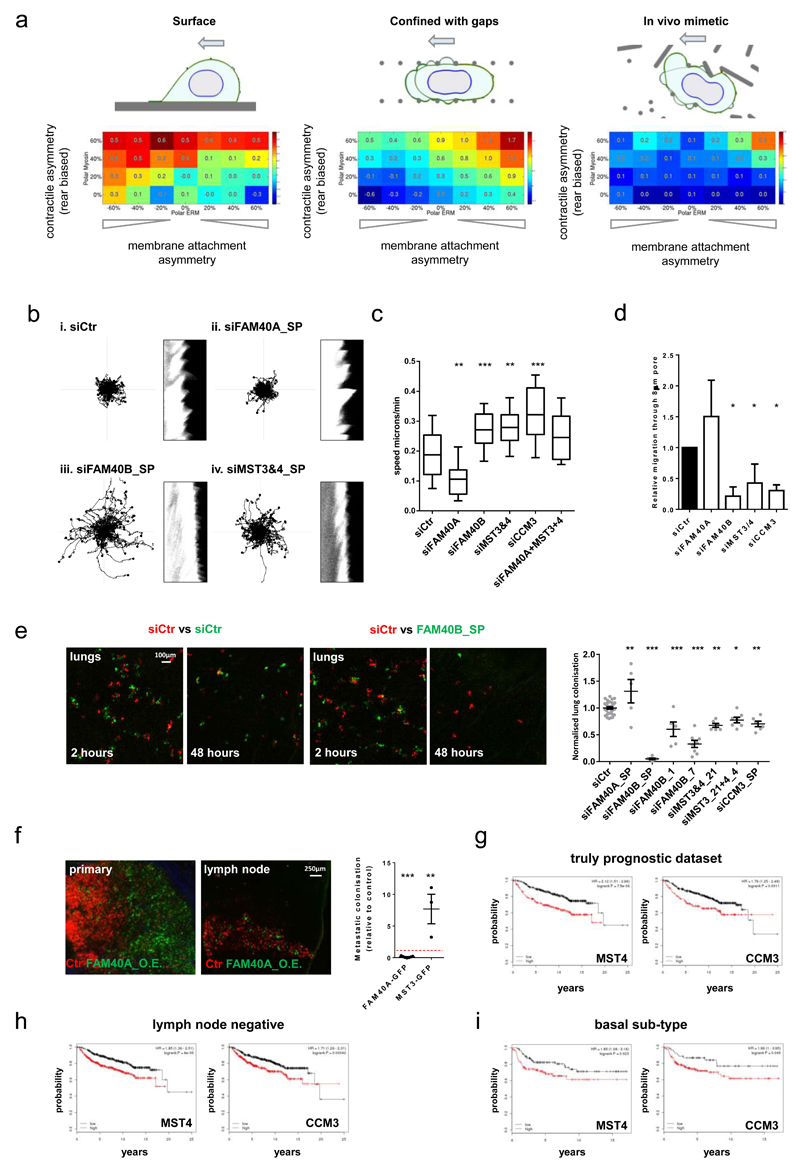
Figure 6. FAM40B, MST3&4, and CCM3 antagonise 2D migration but are required for metastasis.
(a) Matrix geometries investigated using computational model: flat 2D migration, squeezing through a series of gaps, in vivo mimetic environment. (b) 2D analysis of siRNA transfected MDA-MB231 cells (left panels; migration tracks, right panels; kymographs) Kymographs of the cell edge periodicity were generated from high resolution fluorescence images taken over 3minutes at 1second intervals. (c) Analyses of 2D migration speed: Box and whiskers graph: Line=Median, Box=distribution of 50% of values, Whiskers=20-80 percentile. (n=fields of cells (10x); Ctr,34; siFAM40A,37; siFAM40B,30; siMST3&4,35; siCCM3,32; siFAM40A & MST3&4,34). (d) Depletion of FAM40B, MST3&4 and CCM3 prevents MDA-MB231 cell migrating through 8μm pores 6.5µm deep. (n=4 experiments). (e) Mice were injected with mix of Che-expressing control cells and GFP-expressing siRNA-transfected cells. Images show tumour cells lodging the lung parenchyma. The metastatic burden was quantified 2h and 48h post injection by determining the ratio of GFP and Che positive tumour area and normalised to control-injected mice. n as “for Ctr, siFAM40A, siFAM40B-sp, siFAM40B-1, siFAM40B-7, siMST3_21, siMST3&4, siCCM3-sp, n= 31, 5, 5, 5, 8, 6, 7, 6 mice pooled from 7(Ctr), 2 (siFAM40A-sp, siFAM40B-1, siMST_21, siMST3&4, siCCM3-sp) and 3 (siFAM40B-sp, siFAM40B-7) experiments. (f) Mice were injected with a mix of control MDA-MB-231-Che and either MDA-MB-231-FAM40A-GFP or MST3-GFP cells in a ratio (1:1) into the mammary fat pad and the inguinal lymph nodes where imaged after 2-4 months. The spontaneous metastatic burden was quantified by determining the ratio of GFP and Che positive tumour area and normalised to the GFP/Che ratio of the primary tumour (right panel). Images of primary and lymph nodes are shown (right panels). Each mouse is represented in the chart. n=mice; (FAM40AGFP,7; MST3GFP,3). (g-i) Kaplan-Meier plots of distant metastasis free survival of breast cancer patient (n=1809 patients). The analysis includes (g) a truly prognostic dataset, (h) early stage lymph node negative patients (i) and aggressive basal subtype of breast cancer patients. The KM analyses for MST4 and CCM3 were performed using the publicly available datasets http://kmplot.com/analysis/. All statistical test were performed using 1way ANOVA, Sidak’s multiple comparison test, *P<0.05,**P<0.01,***P<0.001. All experiments were conducted at least 3 independent times. Scale bars as indicated.