Figure 4.
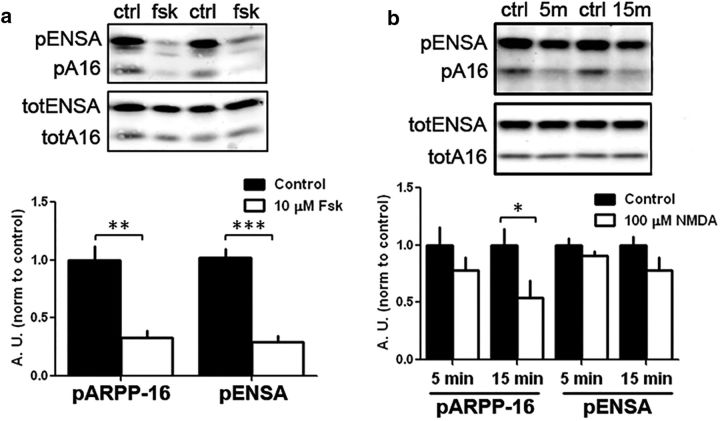
Regulation of phosphorylation of ARPP-16 at Ser46 and ARPP-19/ENSA at Ser62 in striatal slices. a, Rat striatal slices (n = 6 per condition) were treated without (Control, ctrl) or with 10 μm forskolin (fsk) for 10 min. Samples (30 μg total lysate) were analyzed by immunoblotting for phospho-Ser46 and total ARPP-16 or phospho-Ser62 and total ENSA (top). Phospho-site signals were each normalized to total protein, and data for forskolin treatment were normalized to controls (bottom bar graph). **p < 0.01 (Student's t test). ***p < 0.001 (Student's t test). Error bars indicate SEM. b, Striatal slices (n = 3 or 4 per condition) were treated without (ctrl) or with 100 μm NMDA for 5 min (5m), or 15 min (15m), and differences in phosphorylation of ARPP-16 at Ser46 and ENSA at Ser62 were analyzed as in a. *p < 0.05 (multivariate ANOVA with Bonferroni post hoc analysis). Error bars indicate SEM.