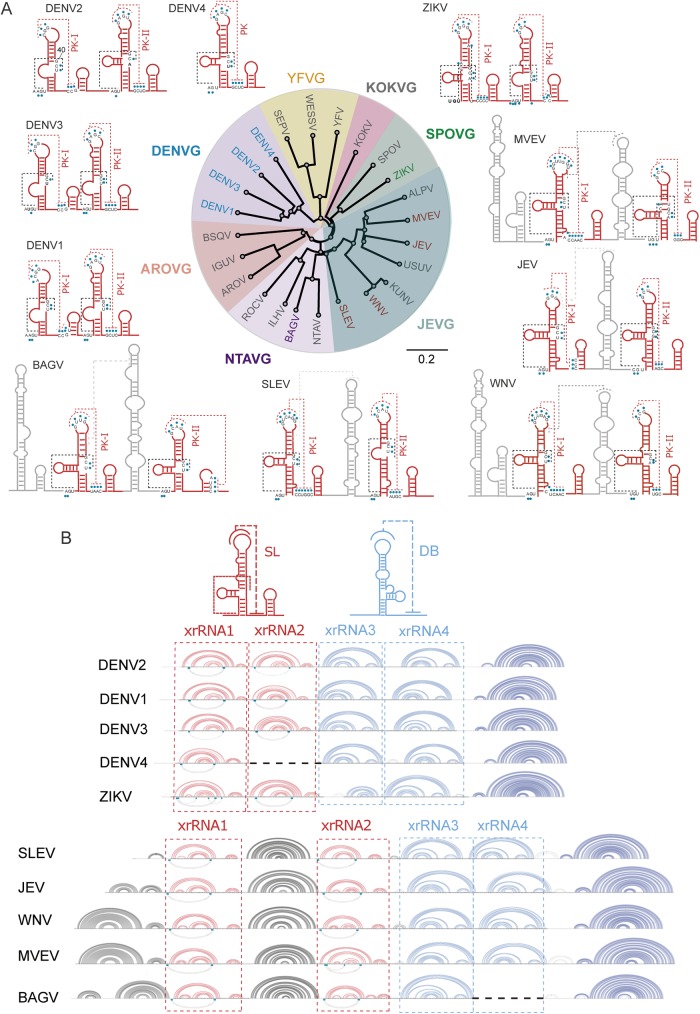
Fig 3. Comparative analysis of predicted RNA structures of the 3’UTR of mosquito-borne flaviviruses (MBFV).
(A) Conserved stem loops of selected viruses from each subgroup of MBFV are shown in red. Predicted H-type pseudoknots are indicated with red dashed lines and pseudoknots including nucleotides present in the three-way junction are shown in black dashed lines. Information obtained from crystallographic studies using MVEV and ZIKV RNAs was included [25,39]. Group-specific RNA structures are indicated in grey. The distance tree was drawn using the neighbor joining method of all complete genome sequences for each virus available in GenBank. (B) Representation of the complete 3’UTR structure of different MBFVs showing the conserved elements involved in Xrn1 stalling. Conserved stem loop (SL) and dumbbell (DB) structures are shown in red and blue, respectively. Arc plots of RNA structures corresponding to the conserved xrRNA1, xrRNA2, xrRNA3 and xrRNA4 are shown for DENV1 to 4, ZIKV, WNV, MVEV, SLEV and BAGV. The conserved 3’SL structure is shown in dark blue.