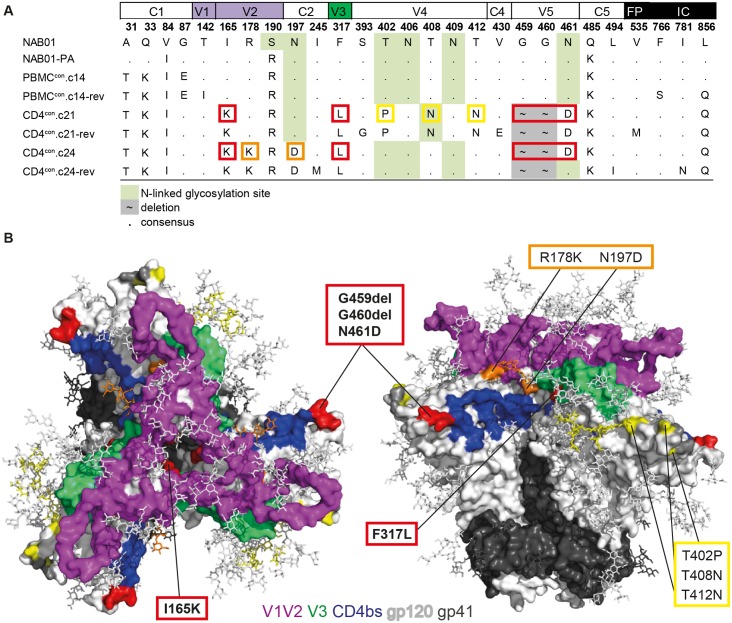
Fig 5. Gp120 sequence mutation pattern following adaptation to CD4low targets.
(A) Summary of amino acid mutations acquired as a result of adaptation to long term in vitro culture and adaptation to CD4low. Green shading indicates mutations affecting N-linked glycosylation sites. Red boxes denote mutations that occur in both CD4low adapted clones, yellow and orange boxes indicate mutations that occurred only in CD4low.c21 and CD4low.c24, respectively. (B) Structural representations of mutated residues in NAB01 associated with adaptation to low levels of CD4 mapped onto crystal structure 5fyj of X1193.c2 SOSIP [100]. Dark gray residue shading indicates limits of non-resolved region of gp120 V4 loop. Missing from the model are the residue at 402, within the non-resolved region of V4, and the glycans at residues 408 and 461 which are not present on this subtype G Env variant. Structure rendered using PyMol version 1.4.1 [101].