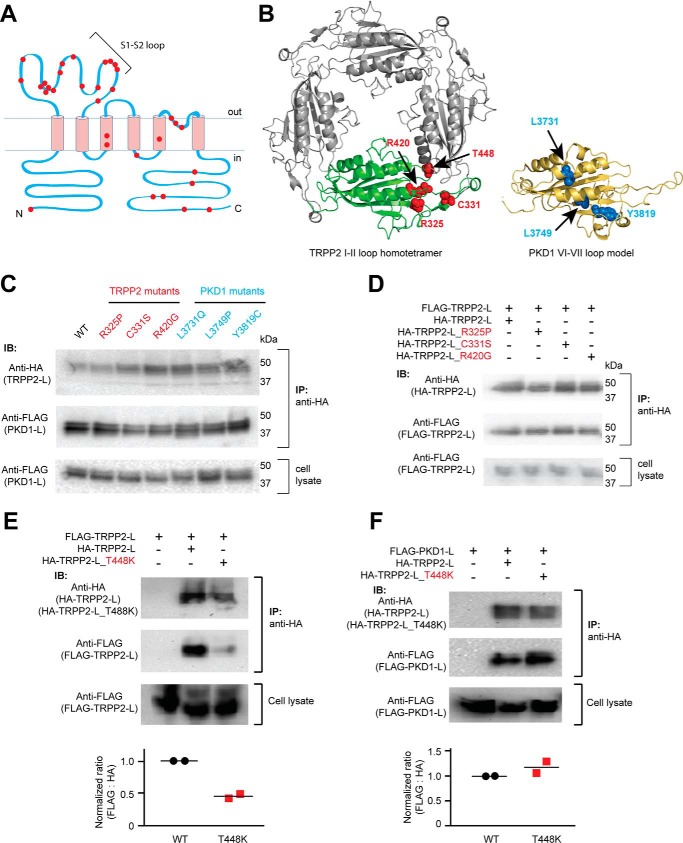
FIGURE 5.
Effects of the ADPKD pathogenic mutations on the assembly of the extracellular loops. A, transmembrane topology of TRPP2 showing that most of the clinically identified single-point mutations (including substitution and deletion, indicated with red dots) are located in the S1-S2 loop. B, localization of the tested mutations in the loop structures. Left, amino acids shown as red van der Waals spheres indicate the four TRPP2 mutations mapped on the TRPP2 S1-S2 loop homotetramer viewing from the top (adapted from the cryo-EM structure of TRPP2, Protein Data Bank code 5T4D (9)). Right, amino acids in blue spheres show the three PKD1 mutations mapped on a structure model of the PKD1 S6-S7 loop made based on the TRPP2 cryo-EM structure. The model was generated on the SWISS-MODEL server (48). Structural graphics were prepared with the program PyMOL (49). C, three TRPP2 mutations in the S1-S2 loop and three PKD1 mutations in the S6-S7 loop had no effect on the assembly between TRPP2 and PKD1 loops. Co-IP was done between WT or mutant loop fragments proteins with anti-HA antibody-coated beads (same for all results in this figure). D, co-IP results indicate that the three mutations have no effect on homomeric assembly of the TRPP2 S1-S2 loop. E, pathogenic mutation T448K significantly weakened the homomeric assembly between TRPP loops. The scatter plot on the bottom shows the normalized ratios of the relative band intensity of the co-immunoprecipitated FLAG-TRPP2 loop to the indicated HA-TRPP2 loops. Data from two independent experiments and the mean (black bars) are shown. F, T448K has no effect on the assembly between TRPP2 and PKD1 loops. The scatter plot shows the normalized ratios of the relative band intensity of the co-immunoprecipitated FLAG-PKD1 loop to the indicated HA-TRPP2 loops. Data from two independent experiments and the mean (black bars) are shown. IB, immunoblotting.