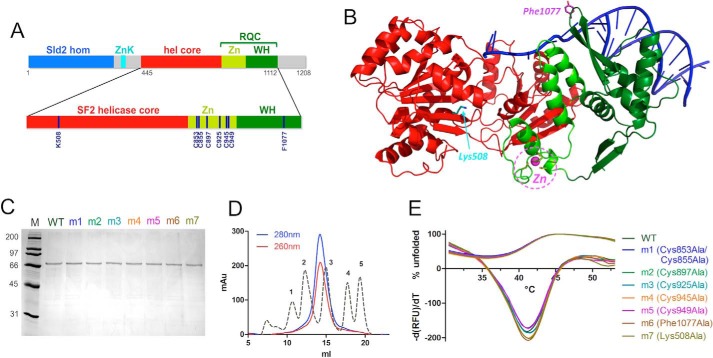
FIGURE 1.
Expression and purification of human RecQ4. A, domain organization of human RecQ4 showing the Sld2 homologous region at the N terminus (blue) followed by a zinc knuckle (cyan), a helicase core (red), and RQC domain (consisting of a zinc-binding domain (bright green) and a winged helix domain (dark green)). Enlarged at the bottom of the panel is the catalytic core of RecQ4 (helicase and RQC domain) with the positions of the residues mutated in this work. B, the crystal structure of RecQ1 (PDB code 2WWY) with the same color code; critical residues are highlighted. C, SDS-PAGE gel of purified proteins. M, molecular markers; m1, C853A/C855A; m2, C897A; m3, C925A; m4, Cs945A; m5, C949A; m6, F1077A; m7, K508A. D, size exclusion chromatography indicates that the recombinant protein is a monomer. The calibration peaks correspond to protein markers: peak 1, ferritin, 440 kDa; peak 2, aldolase, 158 kDa; peak 3, ovalbumin, 44 kDa; peak 4, ribonuclease A, 13.7 kDa; peak 5, aprotinin, 6.5 kDa. E, heat denaturation profiles of RecQ4 variants. The curves depicting the percentage of unfolded proteins (positive y quadrant, 0–100) and the −(dRFU)/dT curves are plotted together (mostly in the negative y quadrant). The experiment suggests that all the mutants are folded and stable.