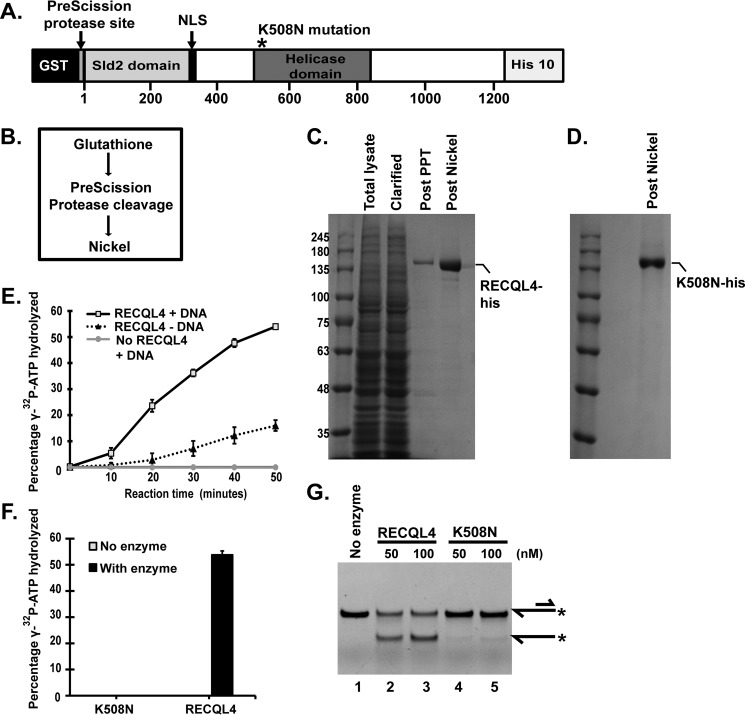
FIGURE 1.
Purification and biochemical analysis of human RECQL4 helicase and K508N-RECQL4 mutant. A, schematic of the recombinant human RECQL4 construct expressed in SF9 cells with a C-terminal His tag and an N-terminal GST tag attached to RECQL4 by the PreScission protease cleavage sequence. * represents position of the K508N mutation for K508N-RECQL4 purification. B, purification strategy for RECQL4 and K508N-RECQL4. C, purification stage fractions of RECQL4 protein. Total lysate, total lysate of protein expressing SF9 cells; Clarified lysate, total lysate supernatant after centrifugation; Post PPT, clarified lysate applied to glutathione column followed by PreScission protease treatment (PPT); Post Nickel, nickel column concentration of the protein in the supernatant of the post PPT fraction. D, final post nickel purification fraction of K508N-RECQL4 (purification columns and process, the same as RECQL4). E, ATPase assay. The plot depicts percentage of [γ-32P]ATP hydrolyzed in the presence of 40 nm RECQL4 and DNA, as well as with no-enzyme and no-DNA controls at indicated time points assayed by TLC. The data points are the mean values of triplicate experiments with S.D. shown as error bars. F, bar graph showing percentages of [γ-32P]ATP hydrolyzed in the presence of 40 nm RECQL4 or the helicase-dead mutant K508N. G, polyacrylamide gel showing DNA helicase activity of RECQL4 and K508N on 5′-FAM-labeled 3′ overhang substrate (20 nm). Substrate and product structures are illustrated on the right side of the gel. * represents the position of FAM label on the substrate.