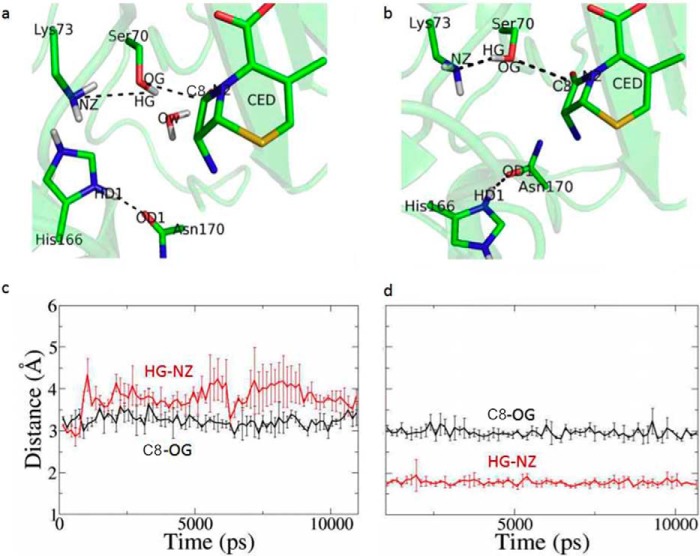
FIGURE 5.
MD simulations to compare two models of the enzyme-substrate complex. Two models of the enzyme-substrate complex were constructed, one with His-166 adopting the “in” conformation as seen in the apo structure and the other with His-166 adopting the “flipped-out” conformation of the ES*-acylation structure. a and c, time course of the distance of Ser-70HG–Lys-73NZ (red) and Ser-70OG-C3 (black) with His-166 adopting “in” conformation. b and d, time course of the distance of Ser-70HG–Lys-73NZ (red) and Ser-70OG-C3 (black) with His-166 adopting flipped-out conformation.