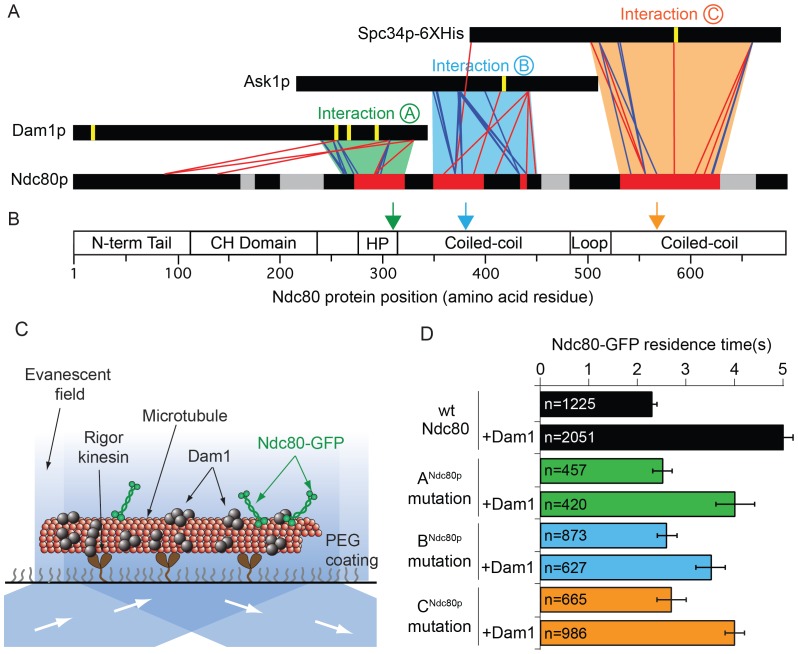
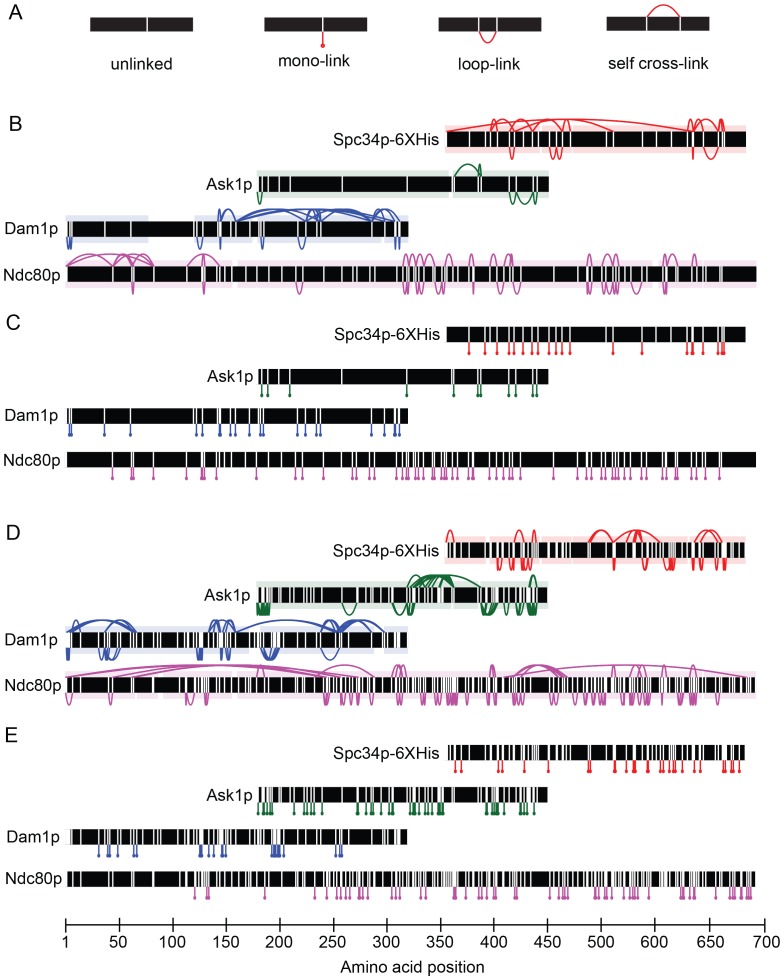
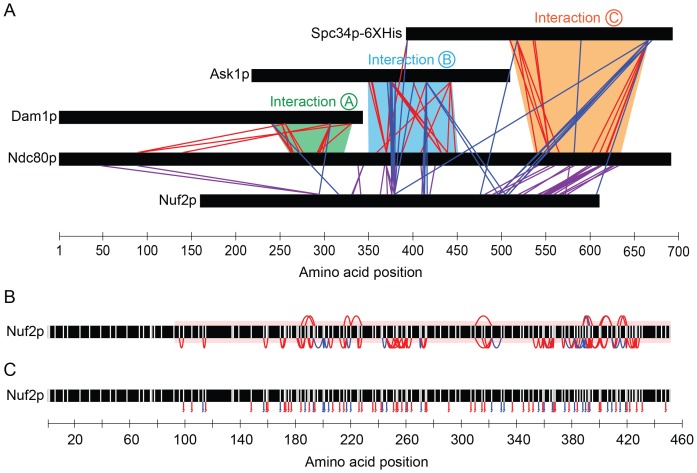
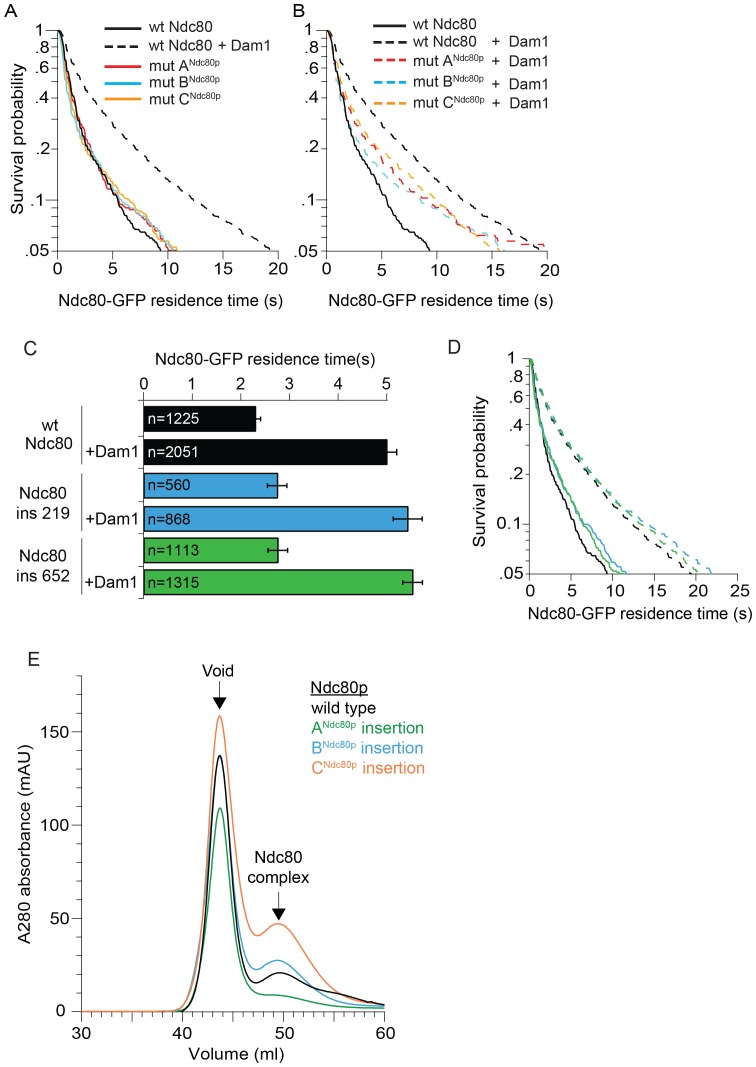
Figure 1. Dam1p, Ask1p, and Spc34p form cross-links to three distinct Ndc80p regions.
(A) Cross-links between the Dam1p, Ask1p, and Spc34p of the Dam1 complex and Ndc80p of the Ndc80 complex. Dam1 and Ndc80 complexes were cross-linked in the presence of microtubules. Horizontal black bars represent proteins and the six vertical yellow lines indicate Aurora B kinase phosphorylation sites on the Dam1 complex. Red and blue lines show cross-links formed with DSS and EDC cross-linkers, respectively. For clarity, only the cross-links between the Dam1 complex proteins and Ndc80p are displayed. Data are shown for peptides with Percolator (Käll et al., 2007) assigned q-values ≤ 0.05. Red bars on Ndc80p indicate regions where clusters of lethal mutations mapped (from Tien et al., 2013) to cross-linked regions. Grey bars on Ndc80p indicate clusters of lethal insertions outside of cross-linked regions. Green, blue, and orange trapezoids represent putative interactions (A, B, and C) between the Dam1 and Ndc80 complexes. (B) Bar diagram of Ndc80p with structural features. Green, blue and orange arrows indicate the positions of lethal mutations in interaction regions ANdc80p, BNdc80p, and CNdc80p used in this study. (CH: calponin homology; HP: hairpin) (C) Diagram showing the setup of TIRF microscopy experiments. Single molecule Ndc80-GFP complex binding on microtubules was visualized in the presence or absence of the Dam1 complex. (D) Lethal mutation in region ANdc80, BNdc80, or CNdc80 partially disrupts the Ndc80 complex’s interaction with Dam1 complex. Average residence time of Ndc80-GFP mutant and wild-type complexes on microtubules in the presence or absence of Dam1 complex. Bars represent average residence time ± error of the mean (estimated by bootstrapping analysis; see Materials and methods for additional details). Ndc80-GFP complex microtubule residence time raw data are included in Figure 1—source data 1. Refer to Supplementary file 1A for statistical analysis of data in part (D).
DOI: http://dx.doi.org/10.7554/eLife.21069.003