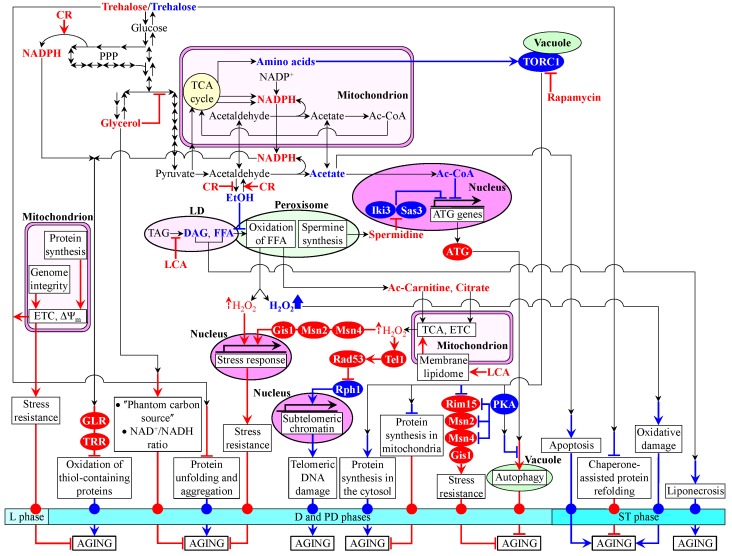
Figure 4. FIGURE 4: A concept of a biomolecular network underlying chronological aging in yeast.
A model for how a stepwise progression of a biomolecular network of cellular aging through a series of lifespan checkpoints is monitored by some checkpoint-specific "master regulator" proteins. The model posits that a synergistic action of these master regulator proteins at several early-life and late-life checkpoints modulates certain vital cellular processes throughout lifespan - thereby orchestrating the development and maintenance of a pro- or anti-aging cellular pattern and, thus, defining longevity of chronologically aging yeast. Activation arrows and inhibition bars denote pro-aging processes (displayed in blue color) or anti-aging processes (displayed in red color). Pro-aging or anti-aging master regulator proteins are displayed in blue color or red color, respectively. Ac-Carnitine, acetyl-carnitine; Ac-CoA, acetyl-CoA; CR, caloric restriction; D, diauxic growth phase; DAG, diacylglycerol; ETC, electron transport chain; EtOH, ethanol; FFA, non-esterified ("free") fatty acids; GLR, glutathione reductase; L, logarithmic growth phase; LCA, lithocholic acid; LD, lipid droplet; PD, post-diauxic growth phase; PKA, protein kinase A; TAG, triacylglycerols; TCA, tricarboxylic acid cycle; ST, stationary growth phase; TORC1, target of rapamycin complex 1; TRR, thioredoxin reductase; ΔΨm, electrochemical potential across the inner mitochondrial membrane.