Figure 8. Src induces non-canonical ARBS that likely interact with CRPC-promoting transcription factors.
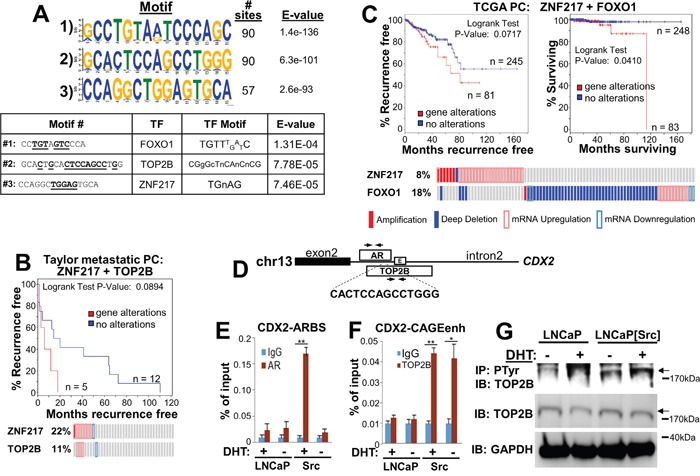
A. Top panel- The three most frequent non-canonical ARBS motifs and their prevalence in the LNCaP[Src527F] AR cistrome analysis. Bottom panel- identification of transcription factor binding site motifs (underlined) within the non-canonical Src-induced ARBS. B. Correlation between ZNF217 and TOP2B differential expression and earlier metastatic recurrence from the study of Taylor et al. [82]. C. Correlation between ZNF217 and FOXO1 differential expression and earlier metastatic recurrence (left panel) and poorer survival (right panel) using PC samples (n = 326) from TCGA (http://cancergenome.nih.gov/). D. The CDX2 Intron 2 contains an AR MACS peak (#3585) from our AR-ChIP-seq analysis of (LNCaP[Src527F] + DHT) cells, encoding the sequence, 5′-CACTCCAGCCTGGG, which is homologous to the non-canonical #2 motif described in panel A. This region also encodes an overlapping TOP2B MACS peak from estradiol-treated MCF-7 (GSE66753), as well as a CAGE enhancer (enh). Arrows, PCR primer sets. ChIP analysis of AR binding to the CDX2-ARBS E. or of TOP2B binding to the CDX2-CAGEenh F. from chromatin prepared from vehicle (-) or DHT (+) treated LNCaP or LNCaP[Src527F] (“Src”) cells. *, p<0.02; **, p<0.001. G. Lysates (1 mg protein/lane) from vehicle (-) or DHT (+) treated LNCaP or LNCaP[Src527F] cells incubated overnight with 4G10(anti-PTyr)-beads were analyzed by IB for TOP2B (arrow). Lysates (20 μg protein/lane) were analyzed by IB for TOP2B or GAPDH. Molecular weight markers are shown at right.
