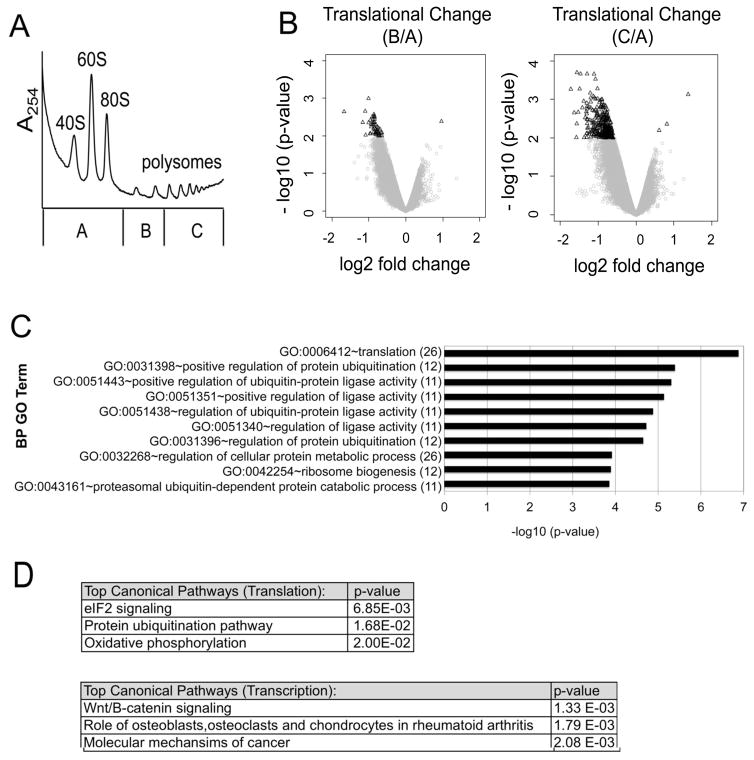
Figure 2. Dexamethasone alters the translation of specific mRNA subsets.
MM1.S cells were treated with 1 μM Dex for 0 or 4 hours. Cell lysates were fractionated by sucrose density ultracentrifugation. (A) For each polysome profile, three distinct gradient fractions (A,B,C) were collected. A representative polysome profile is shown. (B) Volcano plots were generated by comparing translational changes at 4 hours versus 0 hours of Dex treatment. A change in translation was defined as a shift in mRNA transcript distribution between pool B versus pool A (left) or pool C versus pool A (right). The log2 fold change in translation is plotted against the corresponding –log 10 (p-values). A filter of ≤ 0.01 p-value and |log2 fold change| ≥ 0.585 (1.5 fold) was used to define translationally altered genes for each condition (black triangles). To minimize confounding by concomitant transcriptional changes, mRNAs with a |log2 fold change| ≥ 0.585 in transcription were excluded from this analysis. (C) Translationally altered mRNAs from the profiling screen [pool C versus pool A] were subject to gene ontology (GO) analysis and the results are plotted against their corresponding −log 10 (p-values). The number of gene hits in each category is labeled in parenthesis. (D) Ingenuity Pathway Analysis (IPA) was performed to identify the top canonical pathways modulated by Dex at the level of translation versus transcription. The top hits are listed with their corresponding p-values.