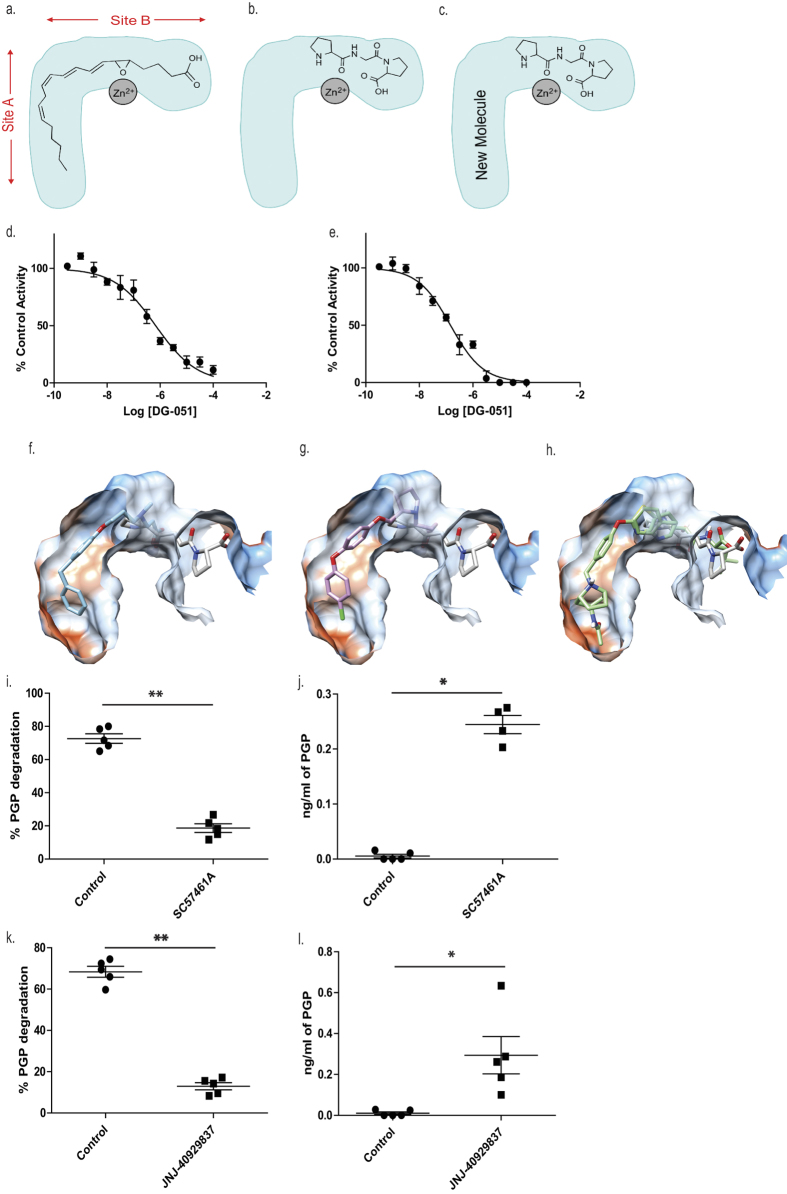
Figure 1. LTA4H inhibitors to have entered the clinic are non-selective and inhibit PGP degradation in vitro and in vivo.
Schematic representation of the binding site of LTA4H showing the relative positions occupied by LTA4 (a) and PGP (b). (c) Proposed solution for a new compound that sits in the narrow arm occupied by the hydrophobic tail of LTA4, (Site A) blocking its hydrolysis, but retaining ability to cleave the tripeptide PGP (Site B). The capacity of DG-051 to inhibit LTA4H epoxide hydrolase (LTB4 generation; (d)) and aminopeptidase activities (PGP degradation; (e)). Overlay of the X-ray structures of ligand-LTA4H structures of (f) SC57461 (blue; 3U9W.pdb) and (g) DG-051 (pink; 3FH7.pdb) with that of the stable PGP analogue OBP-Pro (white, 4MS6.pdb). The protein surface is coloured by amino acid hydrophobicity from most hydrophobic (orange) to hydrophilic (blue) through white regions. (h) JNJ-40929837 (green) was docked into the protein with Autodock Vina. Mice were orally administered SC57461A (i and j) or JNJ-40929837 (k and l) and serum aminopeptidase activity (i and k) or absolute PGP concentrations (j and l) were determined. IC50 curves were generated from 3 technical replicates at each concentration of test compound and at least 2 biological replicates. Data for in vivo studies (i–l) are representative of 2 experiments with 5 mice per group. Results depicted as mean ± SEM. *P < 0.05, **P < 0.01 using Mann–Whitney statistical test.