Fig. 1.
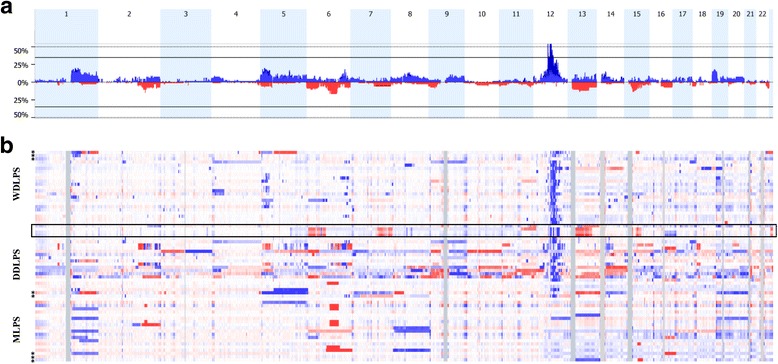
Copy number aberrations (CNAs) identified by array-CGH in 66 LPS samples. a Frequency plot of CNAs in all LPS samples. b Heat map of the CNAs of 66 LPS tumor samples grouped by histological subtypes. Red and blue bars depict percentage of tumors with losses and gains, respectively in the corresponding region of chromosome. X-axis shows the consecutive chromosome numbers. Color intensity on heat map corresponds to the normalized fluorescence log2 ratio from array-CGH experiments. Asterisks (*) indicate samples with the normal genome profile established by conventional karyotyping. Samples denoted by the black frame represent tumors with diagnosis refinement based on array-CGH results
