Abstract
Background
Imported cases of infections due to Dengue (DENV) and Chikungunya (CHIKV) viruses and, more recently, Zika virus (ZIKV) are commonly reported among travelers returning from endemic regions. In areas where potentially competent vectors are present, the risk of autochthonous transmission of these vector-borne pathogens is relatively high. Laboratory surveillance is crucial to rapidly detect imported cases in order to reduce the risk of transmission. This study describes the laboratory activity performed by the National Reference Laboratory for Arboviruses (NRLA) at the Italian National Institute of Health in the period from July 2014 to October 2015.
Methods
Samples from 180 patients visited/hospitalized with a suspected DENV/CHIKV/ZIKV infection were sent to the NRLA from several Italian Hospitals and from Regional Reference Laboratories for Arboviruses, in agreement with the National Plan on human surveillance of vector-borne diseases. Both serological (ELISA IgM test and Plaque Reduction Neutralization Test—PRNT) and molecular assays (Real Time PCR tests, RT-PCR plus nested PCR and sequencing of positive samples) were performed.
Results
DENV infection was the most frequently diagnosed (80 confirmed/probable cases), and all four genotypes were detected. However, an increase in imported CHIKV cases (41 confirmed/probable cases) was observed, along with the detection of the first ZIKV cases (4 confirmed cases), as a consequence of the recent spread of both CHIKV and ZIKV in the Americas.
Conclusions
Main diagnostic issues highlighted in our study are sensitivity limitations of molecular tests, and the importance of PRNT to confirm serological results for differential diagnosis of Arboviruses. The continuous evaluation of diagnostic strategy, and the implementation of laboratories networks involved in surveillance activities is essential to ensure correct diagnosis, and to improve the preparedness for a rapid and proper identification of viral threats.
Background
Vector-borne viral diseases cause a substantial public health burden in tropical and sub-tropical regions. Their geographic distribution is expanding, due to many and complex factors, such as urbanization, climate change, land-use changes, human mobility, and vector range expansion [1].
The Dengue virus (DENV) is a flavivirus (family Flaviviridae) transmitted to humans through Aedes (Ae.) spp mosquito bite. Dengue fever is typically characterized by fever, myalgia, arthralgia, rash, and sometimes severe and life-threatening clinical symptoms. Dengue global incidence has increased 30-fold in the last 50 years [2], and areas with predominant circulation of a single DENV serotype have changed toward co-circulation of different virus serotypes [3]. During the past decade, additional mosquito-borne viruses, including Chikungunya virus (CHIKV) and Zika virus (ZIKV), have successfully spread to geographical areas where only dengue epidemics used to be reported [3–8]. CHIKV is an alphavirus (family Togaviridae) that causes an acute febrile illness characterized by severe arthralgia, whereas ZIKV, another mosquito-borne flavivirus closely related to DENV, mostly causes mild fever, joint pain, conjunctivitis, and rash [3]. As for DENV, CHIKV and ZIKV are transmitted between humans by Ae. species mosquitoes [9, 10]. Since 2004, CHIKV has caused epidemics in Africa, Asia, and Indian Ocean islands. In 2007 an outbreak of chikungunya originated from an imported case coming from India occurred in Italy, causing more than 200 cases of disease [11]. In December 2013, CHIKV was notified in the Caribbean and has since spread to several countries in the Americas [7, 12]. The first outbreak of ZIKV outside Africa and Asia was reported in 2007 in the Yap State, Federated States of Micronesia [13]. Subsequently, in 2013, this virus reappeared in French Polynesia and then spread throughout the Pacific. In the early 2015, the first local transmission of ZIKV was reported in Brazil [14]. Since then, the infection has rapidly spread throughout South America, Central America, and the Caribbean [8, 15, 16], and recently in Florida, USA [17]. ZIKV, previously thought to be associated with a mild clinical disease, was found to be associated with a 20-fold increase in the Guillain-Barrè syndrome incidence following the French Polynesia outbreak [18]. Moreover, the report of a possible association between ZIKV infection and an epidemic of microcephaly among neonates in Brazil has attracted global attention, and has led the World Health Organization (WHO) to declare the ZIKV epidemic as a global public health emergency on February, 1st 2016 [19]. In the meanwhile, evidence supporting the association between ZIKV infection and neonatal microcephaly and other birth defects has increased [20–25].
Imported cases of illness due to DENV and CHIKV, and more recently ZIKV, are reported every year among travelers returning from endemic regions [26–28]. In areas where competent vectors are present, the risk of autochthonous transmission of these vector-borne pathogens is particularly high [11, 29]. Thus, epidemiological and laboratory surveillance is crucial to rapidly identify imported cases in order to introduce measures to reduce risks for public health. The aim of the present study is to present data on imported infections in Italy, diagnosed at the National Reference Laboratory for Arboviruses (NRLA) in the period from July 2014 to October 2015, mainly focusing on diagnostic issues, countries of origin of the infections, and viral strains involved in the imported cases.
Methods
Patients and samples
Samples of patients visited/hospitalized with a suspected DENV/CHIKV/ZIKV infection, collected from July 2014 through October 2015, were analyzed. A case-report form containing information about age, sex, countries visited, travel dates, and date of onset of symptoms was completed for each patient. Samples were collected and sent to the NRLA at the Istituto Superiore di Sanità in Rome from several Italian Hospitals, in agreement with the National Plan on human surveillance of imported and autochthonous vector-borne diseases (CHIKV, DENV, ZIKV, and West Nile virus—WNV) [30, 31]. Samples were sent also from Regional Reference Laboratories for Arboviruses involved in the surveillance National Plan for diagnostic confirmation and/or with the aim of a cross-evaluation of the diagnostic methods used in different laboratories. ELISA IgM and real time PCR tests were performed for a first line diagnosis. Plaque Reduction Neutralization Tests (PRNTs) were performed to confirm positive results obtained by ELISA tests, and to discriminate between closely related viruses.
Serological assays
ELISA IgM
IgM antibodies against DENV, CHIKV, and ZIKV were detected in patients serum samples using commercial IgM-capture ELISA systems (Focus Diagnostics Dengue Virus IgM Capture, DxSelect™, California, USA, NovaLisa® Chikungunya IgM μ-capture ELISA, NovaTec Immundiagnostica GmbH, Germany, Euroimmun Anti-Zika Virus IgM ELISA, Luebeck, Germany). Absorbance was measured at 450 nm using an ELISA reader, according to manufacturer’s instructions. Sample optical density readings were compared with reference cut-off OD readings to determine results. Index values >1.00 for DENV, > 11.00 for CHIKV, and > 1,1 for ZIKV were considered presumptive for the presence of IgM antibodies.
Plaque Reduction Neutralization test (PRNT)
The assay was performed in six-well tissue culture plates with subconfluent VERO cell monolayers (approximately 70% confluence). The following viruses were used: serotype 2 DENV (NGB strain), a CHIKV strain isolated from a patient during the 2007 Italian outbreak [11], and ZIKV H/PF/2013 strain of the Asian genotype (kindly provided by Dr. Isabelle Leparc-Goffart of the French National Reference Center on Arboviruses in Marseille) [32]. Infectivity titration of each viral strain was performed by plaque assay using VERO cells. Patients sera were diluted 1:10 in serum-free maintenance medium, heat-inactivated, and titrated in duplicate in twofold dilution steps. Equal volumes (100 μl) of DENV/CHIKV/ZIKV dilution containing approximately 80 Plaque Forming Units (PFU), and serum dilutions, were mixed, and incubated overnight at 4 °C. Subsequently, VERO cells plates were infected with 200 μl/well of virus-serum mixtures in duplicate. After 1 h incubation at 37 °C and 5% CO2, the inocula were aspirated and the wells were overlayed with a mixture of one part 2% Gum Tragacanth and one part of supplemented medium (2× MEM, 2.5% inactivated FCS and 2% 1 M HEPES). The plates were incubated at 37 °C and 5% CO2 for 2 (CHIKV) - 7 (DENV) - 4 (ZIKV) days, and then were stained with 1.5% crystal violet. A titration of CHIK/DEN/ZIK viruses with three dilutions in duplicate (the working dilution, 1:2 and 1:8 dilutions) was performed in each assay and used as a control for the assay. Neutralizing antibody titers were calculated as the reciprocal of the serum dilution that gave a 50 or 80% reduction of the number of plaques (PRNT50/PRNT80), as compared to the virus control. PRNT80 ≥ 10 were considered positive, while PRNT50 ≥ 10 were considered as border line (b.l.).
Molecular diagnosis
RNA extraction and Real Time PCRs
Molecular tests were performed on acute sera of DENV/CHIKV/ZIKV-suspected patients.Viral RNA was extracted from 140 μl of serum sample by using QIAmp viral RNA Mini kit (Qiagen Inc., Valencia, CA, USA), according to the manufacturer’s instructions, and then stored at -80 °C until further processing. The RNA was amplified by real time PCR for CHIKV, DENV, and/or ZIKV detection. The primers and probes used in this study are listed in Table 1 [11, 33, 34]. All real time PCR assays were performed by using the RealTime ready RNA Virus Master mix (Roche Diagnostics, Basel, CH), according to the manufacturer’s protocol, and CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad).
Table 1.
Primers and probes for the molecular diagnosis
| Primers and Probes | Sequence (5’- 3’) | Reference |
|---|---|---|
| DenS | GGATAGACCAGAGATCCTGCTGT | [33] |
| DenAs + DenAs1 | CATTCCATTTTCTGGCGTTC + CAATCCATCTTGCGGCGCTC | |
| DenP | FAM-CAGCATCATTCCAGGCACAG-TAMRA | |
| ChikS | TGATCCCGACTCAACCATCCT | [11] |
| ChikAs | GGCAAACGCAGTGGTACTTCCT | |
| ChikP | FAM-TCCGACATCATCCTCCTTGCTGGC-Black Hole Quencher 1 | |
| Zvf1086 | CCGCTGCCCAACACAAG | [34] |
| Zvr1162c | CCACTAACGTTCTTTTGCAGACAT | |
| ZvP_1107 | 6FAM-AGCCTACCTTGACAAGCAGTCAGACACTCAA-TAMRA |
Amplification and sequencing from viral RNA
For DENV and CHIKV nucleic acid detection and genotyping, Reverse Transcription (RT)-PCR followed by nested PCR amplification was performed. An amplicon of 434 bp in the E gene region and an amplicon of 536 bp in the E1 structural glycoprotein coding gene region were obtained for DENV an CHIKV, respectively. The primers used for RT-PCR plus nested PCR and sequencing are listed in Table 2 [35, 36]. SuperScript One-step RT-PCR with Platinum Taq kit (Invitrogen, Gaithersburg, MD) and Platinum PCR SuperMix kit (Invitrogen) were used for RT-PCR and nested PCR, respectively. PCR products were purified by QIAquick PCR Purification Kit (Qiagen) and were sequenced on both strands by using nested forward and reverse primers.
Table 2.
Primers for DENV and CHIKV amplification and sequencing
| Primers | Sequence (5’- 3’) | Reference |
|---|---|---|
| RT-PCR: | [35] | |
| DEUL | TGGCTGGTGCACAGACAATGGTT | |
| DEUR | GCTGTGTCACCCAGAATGGCCAT | |
| Nested PCR: | ||
| DENUL | GATCTCAAGAAGGAGCCATGCA | |
| DENUR | ATGGAACTTCCCTTCTTGAACCA | |
| RT-PCR: | [36] | |
| CHIK 10264 F | GGCGCCTACTGCTTCTG | |
| CHIK 11300R | CGACACGCATAGCACCSC | |
| Nested PCR: | ||
| CHIK 10564 F | CCCTTTGGCGCAGGAAGAC | |
| CHIK 11081R | GACTTGTACGCGGAATTCGG | |
Phylogenetic analysis
The sequences obtained were aligned with other DENV/CHIKV sequences available in the GenBank database (accession numbers are reported in the phylogenetic trees), by using the ClustalW program (www.clustal.org) [37]. Alignments were manually edited with the Bioedit program [38]. Nucleotide Tamura-Nei model and the Neighbour-Joining method was used to construct the phylogenetic trees [39]. The Neighbour-Joining method was implemented by using MEGA version 6.06 (www.megasoftware.net) [40]. The robustness of branching patterns was confirmed with a bootstrap analysis using 1000 replicates.
Results
Dengue, Chikungunya and Zika diagnostic tests results, and areas of origin of the imported infections
Samples collected from 180 patients visited/hospitalized with a suspected DENV/CHIKV/ZIKV infection were analyzed. Of the patients, 50,6% were males, median age was 38 years (range 1–80 years). For 116 patients for whom the information about the date of symptoms onset and/or hospitalization was available, the median lag time before sample collection was 8 days (range 2–102 days, mean ± standard deviation: 15,47 ± 18,24 days). Two serum samples (acute phase and convalescence phase) were available from 27 patients. Samples were sent to the NRLA from several Italian Regions (Friuli Venezia Giulia, Lombardia, Piemonte, Liguria, Toscana, Umbria, Lazio, Abruzzo, Campania, Sardegna, Calabria e Sicilia). Most of the samples were collected during the summertime, from June to September, when the surveillance is increased because of vector activity.
On the base of diagnostic tests results, and clinical and epidemiological data, each case was defined as confirmed, probable, possible or not confirmed, according to the criteria shown in Table 3.
Table 3.
Case definition on the bases of the diagnostic test results
| Confirmed | PCR positive and/or IgM positive plus PRNT positivea, and/or seroconversion or four fold increase in neutralizing antibody titers in two consecutive samples. |
| Probable | IgM positive plus PRNT border lineb in acute samplesc. |
| Possible | IgM negative and PRNT positive/border line, or IgM positive but PRNT negative in acute samples. |
| Not confirmed | IgM positive and PRNT negative in late/convalescent samples, or PRNT positive without an increase in the titer in two consecutive samples. |
aPRNT80 ≥ 10: positive
bPRNT50 ≥ 10: border line (b.l.)
cThese cases were classified as possible if PRNT b.l. results were obtained toward different viruses
In the study period, a total of 157 patients were tested for DENV, 97 for CHIKV and 16 for ZIKV (Table 4).
Table 4.
DENV, CHIKV and ZIKV diagnosis in the period from July 2014 to October 2015
| Total | Confirmed | Probable | Possible | Not confirmed | |
|---|---|---|---|---|---|
| DENV diagnosis | 157 | 68 | 12 | 33 | 44 |
| CHIKV diagnosis | 97 | 35 | 6 | 14 | 42 |
| ZIKV diagnosis | 16 | 4a | 0 | 3b | 9 |
| Dual diagnosis DENV/CHIKV | 76: two cases of possible co-infections. | DENV: 18/76 (of which two CHIKV confirmed and one CHIKV possible cases) | |||
| CHIK: 20/76 (of which two DENV confirmed and 6 DENV possible cases) |
aThe diagnosis of one of these cases was performed in Germany after we excluded DENV and CHIKV infections (ref). One was a case of autochthonous (most likely sexual) transmission (ref)
bOf these, two were probable cases of past ZIKV infections (PRNT positives and IgM negatives). One showed instead a PRNT b.l. result for ZIKV, which was probably due to cross reactivity of DENV specific antibodies
Overall, 68 DENV cases (plus 12 probable cases), 35 CHIKV cases (plus 6 probable cases), and 4 ZIKV cases [27, 41] were confirmed, plus two cases of ZIKV past infection. For 76 patients, diagnostic tests both for DENV and for CHIKV were performed, and 2 cases of possible co-infections were detected. Clinical features of DENV, CHIKV and ZIKV confirmed/probable cases are shown in Table 5.
Table 5.
Clinical features of DENV, CHIKV and ZIKV confirmed/probable cases
| Symptoms | DENV confirmed and probable cases presenting with the symptoma | CHIKV confirmed and probable cases presenting with the symptoma | ZIKV confirmed and probable cases presenting with the symptoma |
|---|---|---|---|
| Fever (≥38 °C) | 93,2% | 92,6% | 75,0% |
| Arthralgia | 71,2% | 96,3% | 75,0% |
| Rash | 33,9% | 66,7% | 100,0% |
| Asthenia | 76,3% | 70,4% | 25,0% |
| Headache | 62,7% | 37,0% | 0,0% |
| Myalgia | 52,6% | 63,0% | 25,0% |
| Retro-orbital pain | 32,2% | 11,1% | 0,0% |
| Meningoencephalitis | 1,7% | 3,7% | 0,0% |
| Others | 10,2%b | 3,7% c | 0,0% |
aSymptoms were known for 59, 27 and 4 DENV, CHIKV and ZIKVV cases, respectively
bdiarrhea, vomit, leuco-thrombocytopenia
csymptoms persisting for longer than 30 days
The area of origin of the suspected imported cases was known for 62 DENV, 30 CHIKV, and 4 ZIKV confirmed/probable cases DENV cases were imported from many different countries in all continents except Europe. Among DENV confirmed/probable cases, 59.7% were from Asia, 17.7% from Central and South America, 11.3% from the Caribbean, 6.5% from Africa, and 4.8% from Oceania. As expected, because of the recent CHIKV epidemics in the Americas, among the CHIKV confirmed/probable cases, 53.3% were from the Caribbean, 36.7% were from Central and South America, and only 10.0% were from Asia or Africa. Two ZIKV confirmed infections had been acquired in Brazil, one in March [27] and one in May, 2015, while one had been acquired in Thailand in 2014, and one was an autochthonous case likely due to sexual transmission [41].
Serological diagnosis of DENV, CHIKV and ZIKV infections
ELISA IgM tests
Results of ELISA IgM tests are summarized in Table 6.
Table 6.
DENV/CHIKV/ZIKV ELISA IgM tests results
| ELISA IgM: positives/tested | Estimated proportion of false positive and false negative test results | |
|---|---|---|
| DENV (Focus Diagnostics Dengue Virus IgM Capture, DxSelect™) | 55/127 | 8/55 (14.5%) false positives |
| 8/72 (11.1%) false negatives | ||
| CHIKV (NovaLisa® Chikungunya IgM μ-capture ELISA, NovaTec Immundiagnostica) | 31 + 3 b.l./86 | 1 b.l./86 (1,2%) false positives |
| 5/52 (9,6%) false negatives | ||
| ZIKV (Euroimmun IgM ELISA) | 3/5 |
DENV ELISA IgM test was performed for 127 of 157 patients tested for DENV, with 55 positive results; for 14.5% of the ELISA IgM positive patients, a final diagnosis of confirmed or probable DENV infection was not done after considering all laboratory findings and available epidemiological data. These cases are likely to represent false positive IgM ELISA results. Conversely, 11.1% of the ELISA IgM negative patients were diagnosed as confirmed DENV cases on the basis of other tests (PCR positivity) and/or of IgM results obtained by the hospital/laboratory where the sample had been collected (not shown); they were considered as false negatives. Of note, of the DENV suspected cases coming to our laboratory with a positive IgM result obtained in the hospital/laboratory where the sample had been collected (n = 53) (not shown), 24.5% (13/53) could not be confirmed by PRNT nor by molecular tests.
CHIKV ELISA IgM test was performed for 86 of 97 patients tested for CHIKV, with 3 b.l. results, and 31 positive results; all positive ELISA IgM results and two of the b.l. were confirmed by a positive or b.l. PRNT result. Among 52 CHIKV ELISA IgM negative samples, 13 were positive and 4 b.l. in PRNT: of these, at least 5 (9.6%) were considered to be associated with a recently acquired infection, based on clinical and epidemiological data, and thus estimated as probable false-negative ELISA results. Of the CHIKV suspected cases coming to our laboratory with a positive IgM result obtained in the hospital/laboratory where the sample had been collected (n = 27) (not shown), 11.1% (3/27) were classified as not confirmed after evaluation of all laboratory findings. ZIKV IgM test was performed for 5 patients, all with a PRNT positive result for ZIKV: 3 were positive, and were thus considered as recent, confirmed, ZIKV infections, while the two IgM negatives were considered as past infections.
Overall, 94 DENV and 40 CHIKV ELISA IgM results could be compared with the IgM results obtained with different methods in the hospital/laboratory of origin of the samples (not shown): concordant results were obtained in 81.9 and 87.5% of cases for DENV and CHIKV, respectively.
PRNTs
PRNTs results are summarized in Table 7: neutralizing antibodies were detected in 79/157 (50.3%) of patients tested for DENV, in 47/97 (48.4%) of patients tested for CHIKV, and in 5/15 (33,3%) of patients tested for ZIKV. In 26/79 (32,9%) of DENV PRNT positive patients, 10/47 (21,2%) of CHIKV PRNT positive patients, and 2/5 (40%) of ZIKV PRNT positive patients, both molecular tests and ELISA IgM gave negative results: these subjects had probably acquired a DENV and/or CHIKV infection in the past, which was not associated with the recent/ongoing illness. A b.l. PRNT result was obtained for 31 of 157 DENV tested patients. Of these, 10 (32.3%) were classified as confirmed cases, since the viral genome could be detected in the same sample, and/or a fully positive PRNT result was obtained in a second, convalescent sample. Moreover, 12 (38.7%) were classified as probable cases, since a positive ELISA IgM results was obtained in the same sample. Finally, 9 (29%) PRNT b.l. results were obtained from cases defined as possible, which were not associated with any other positive result (n = 5), and/or associated with a confirmed infection by a closely related Flavivirus (ZIKV, n = 3), and/or in cases showing b.l. PRNT results also for different viruses (such as CHIKV and WNV, n = 3), suggesting a broad and non DENV-specific cross-reactivity. With respect to CHIKV diagnosis, a b.l. PRNT result was obtained for 11 (11.3%) of 97 CHIKV tested patients: one from a confirmed case, 4 from probable cases, and 6 which were classified as possible cases, since they were not associated with positive results in other CHIKV tests, and, in 3 cases, presented b.l. PRNT results also for different viruses (such as DENV and WNV). Finally, one b.l. PRNT result was obtained for ZIKV, in a sample of a DENV confirmed case.
Table 7.
DENV/CHIKV/ZIKV PRNTs results
| PRNT positives/tested | PRNT border line/tested | ||
|---|---|---|---|
| DENV | 79/157 (50.3%) | 31/157 (19,7%) | 10/31 (32.3%): confirmed cases |
| 12/31 (38.7%): probable cases | |||
| 9/31 (29%): possible cases | |||
| CHIKV | 47/97 (48.4%) | 11/97 (11,3%) | 1/11 (9%): confirmed case |
| 4/11 (36,4%): probable cases | |||
| 6/11 (54,6%): possible cases | |||
| ZIKV | 5/15 (33,3%) | 1/15 (6,7%) | possible case (probable cross reactivity of DENV specific neutralizing antibodies) |
Molecular diagnosis of DENV and CHIKV infections, and phylogenetic analysis of viral sequences
For DENV diagnosis, 25 of 132 (18.9%) samples tested by real time PCR gave a positive result. All PCR positive samples (for which the time from the onset of symptoms was known), had been collected within 8 days from the onset of symptoms (mean ± standard deviation: 4.71 ± 1.76 days). For CHIKV diagnosis, only 2/76 (2.6%) samples tested by real time PCR gave a positive result, which had been collected 3 days after the onset of symptoms. All samples analyzed for ZIKV by real time PCR gave negative results.
Among all samples collected within 8 days from the onset of symptoms, CHIKV viral genome was detected in 7.7% (2/26) of the samples (22.2% of confirmed/probable cases), and DENV viral genome in 36.8% (21/57) of tested samples (53.8% of confirmed/probable cases). These data may suggest a longer duration of viremia in DENV infection compared to CHIKV infection.
Sequences were obtained for 22 of the DENV PCR positive samples, and for the 2 CHIKV PCR positive samples. Both nucleic acid and translated amino acid sequences were aligned with GenBank sequences of isolates with known dates and locations, and phylogenetic analysis was performed. Viral strains and genotypes, locations of origin, year of the infections, and Gene Bank accession numbers are summarized in Tables 8 (sequences characterized in this study) and Table 9 (reference sequences). As shown in Fig. 1a,b,c and d, DENV strains of all the four serotypes were identified (11 DENV-1, 8 DENV-2, two DENV-3 and one DENV-4 strains). Within the DENV-1 serotype, 4 strains grouped together with the Asian lineage (genotype I), 3 with the South Pacific lineage (Genotype IV) and 4 with African/American lineage (Genotype V), with bootstrap values ≥ 97 (Fig. 1a). Most of DENV-1 sequences (from patients S2015-423, S2015-475, S2014-376, S2015-510, S2015-470, S2015-425, S2015-481, S2015-458, S2015-431) showed a high similarity with GenBank strains known to circulate in the areas of origin of the imported infections (88–98% homology at nucleic acid level and 100% at amino acid level). In contrast, two of our DENV-1 sequences showed a higher degree of divergence when compared to DENV-1 sequences available in Gene Bank. A strain from Philippines, identified in the patient S2014-383, showed the highest similarity (88% homology at nucleic acid level and 93% at amino acid level), with two strains from Philippines collected in previous years (acc. n° JN415517 collected in 2010; acc. n° KR919819 collected in 2012). Moreover, the phylogenetic analysis of the strain S2014-358 from Thailand, showed the higher similarity (86% homology at nucleic acid level and 92% at amino acid level) with a Gene Bank Thai strain collected in 2010 (acc. n° JN415528). The strain S2014-358 showed the same 92% similarity at amino acid level also with other Thai strains collected in 2013 (acc. n° KJ545455 and KF887994). In Fig. 1b is reported the DENV-2 tree: 7 out of 8 strains (S2014-368, S2015-409, S2015-465, S2014-482, S2015-477, S2014-478 and S2015-512) clustered in the Cosmopolitan genotype: 4 of them (S2015-409, S2015-465, S2015-478, S2015-482), from Thailand and India, showed a high similarity with Indian isolates sequences (2001-DQ448236 and 2011-KF364514) both at nucleic acid and amino acid level (90–98% and 100% respectively). The S2014-382 sequence, from Santo Domingo, clustered in the American/Asian genotype, showing a strong homology (85 and 100% in nucleic and amino acid composition, respectively) with the strain collected in Puerto Rico in 2013 (acc. n° LN870427). The two DENV-3 strains identified in this study (S2014-339 from Cuba, and S2015-517 from unknown geographic area) clustered in genotype III, showing 93–100% of identity with the GenBank strains collected in Central American and Caribbean areas (acc. n° DQ341204 and level (Fig. 1c). The DENV-4 sequence (S2015-466, from Thailand) showed a strong homology with an isolate collected in 2013 from Myanmar area (acc. n° KJ470765) (similarity of 90 and 100% at nucleic and amino acid level, respectively), and distance values of 73–80% at nucleic acid level and 83–93% at amino acid level, with other sequences of different years from Thailand (acc. n° AY618990 collected in 1991, AY618980 collected in 1998, AY618992 collected in 2001, and EU448454 collected in 2007) (Fig. 1d). CHIKV sequences obtained from patients S2015-416 (for which the geographic area of origin of the infection was not known) and S2015-422, from Colombia, were aligned with 30 GenBank sequences with known dates and locations. From the analysis of the phylogenetic tree, the two sequences were shown to belong to the Asian genotype (Fig. 2). The S2015-416 sequence showed a 100% identity both at nucleic and amino acid level with an isolate collected in 2008 in Indonesia (acc. n° KC879577). The S2015-422 sequence showed the strongest homology with a Brazilian isolate (acc. n° KP164572) (98 and 96% similarity at nucleic and amino acid level, respectively).
Table 8.
DENV and CHIKV sequences characterized in this study
| Isolate ID | Travel location | Genotype | Year isolated | GenBank accession no. |
|---|---|---|---|---|
| DENV-1 | ||||
| S2014-358 | Thailand | I-Asian | 2014 | LN870423 |
| S2014-376 | Bali, Indonesia | I-Asian | 2014 | LN870425 |
| S2015-510 | ? | I-Asian | 2015 | LN999960 |
| S2015-460 | French_Polynesia | I-Asian | 2015 | LN999954 |
| S2015-475 | Philippines | IV-South Pacific | 2015 | LN999955 |
| S2015-423 | Oceania | IV-South Pacific | 2015 | LN879497 |
| S2014-383 | Manila, Philippines | IV-South Pacific | 2014 | LN870426 |
| S2015-431 | Maldives | V-African/American | 2015 | LN879498 |
| S2015-458 | Maldives | V-African/American | 2015 | LN999951 |
| S2015-481 | Mexico | V-African/American | 2015 | LN999958 |
| S2015-425 | Haiti | V-African/American | 2015 | LN879499 |
| DENV-2 | ||||
| S2014-368 | ? | Cosmopolitan | 2014 | LN870428 |
| S2015-477 | Maldives | Cosmopolitan | 2015 | LN999956 |
| S2015-512 | ? | Cosmopolitan | 2015 | LN999961 |
| S2015-409 | Thailand | Cosmopolitan | 2015 | LN999950 |
| S2015-465 | Thailand | Cosmopolitan | 2015 | LN999952 |
| S2015-482 | India | Cosmopolitan | 2015 | LN999959 |
| S2015-478 | Thailand and Cambodia | Cosmopolitan | 2015 | LN999957 |
| S2014-382 | Santo Domingo, Dominican Republic | America/Asian | 2014 | LN870427 |
| DENV-3 | ||||
| S2015-517 | ? | III | 2015 | LN999962 |
| S2014-339 | Cuba | III | 2014 | LN870424 |
| DENV-4 | ||||
| S2015-466 | Thailand | I | 2015 | LN99995 |
| CHIKV | ||||
| S2015-416 | ? | Asian | 2015 | LN879501 |
| S2015-422 | Colombia | Asian | 2015 | LN879500 |
Table 9.
DENV and CHIKV reference sequences
| Virus strain | Location | Genotype | Year isolated | GenBank accession no. |
|---|---|---|---|---|
| DENV-1 | ||||
| NC14-17042014-4554 | New Caledonia | I-Asian | 2014 | KM212960 |
| China/GD-D13001 | Thailand | I-Asian | 2013 | KJ545455 |
| DENV-1/8/Thailand/01/2013 | Thailand | I-Asian | 2013 | KF887994 |
| Khabar 2759 | Khabarovsk, Far East, Russia | I-Asian | 2012 | KJ417841 |
| SL_2012_GS0319 | Sri-Lanka | I-Asian | 2012 | KJ26662 |
| MKS-WS81 | Indonesia | I-Asian | 2010 | KC762639 |
| D1/Vietnam/1012aTw | Viet Nam | I-Asian | 2010 | JF967953 |
| Thailand 2010 | Thailand | I-Asian | 2010 | JN415528 |
| D1/IDN/Bali_033/2010 | Indonesia | I-Asian | 2010 | KM216676 |
| - | Cambodia | I-Asian | 1998 | AF309641 |
| GZ/80 | China | I-Asian | 1980 | AF350498 |
| PUO 359 | Thailand | I-Asian | 1980 | AF425630 |
| 16007 | Thailand | II-Thailand | 1964 | AF180817 |
| TH-SMAN | Thailand | II-Thailand | 1954 | D10513 |
| P72-1244 | Malaysia | III-sylvatic | 1972 | EF457905 |
| D1/Hu/Philippines/NIID13/2016 | Philippines | IV-South Pacific | 2016 | LC128301 |
| FI/DB170/2014 | Fiji | IV-South Pacific | 2014 | KM279390 |
| Phil2012 | Philippines | IV-South Pacific | 2012 | KR919819 |
| Philippines 2010 | Philippines | IV-South Pacific | 2010 | JN415517 |
| WS01/190801-769 | Samoa | IV-South Pacific | 2001 | JQ655095 |
| A88 | Indonesia | IV-South Pacific | 1988 | AB074761 |
| AUS HCS1 | Australia | IV-South Pacific | 1983 | AF425611 |
| PRS 228682 | Philippines | IV-South Pacific | 1974 | AF425627 |
| Guangzhou/2014/4 | China | V-African/American | 2015 | KT751343 |
| Wenzhou-Human-1 | China | V-African/American | 2014 | KR024708 |
| 9/D1/Del/2013 | India | V-African/American | 2013 | KU166895 |
| AO/DB132/2013 | Angola | V-African/American | 2013 | KM277610 |
| DENV-1/NI/BID-V7696/2012 | Nicaragua | V-African/American | 2012 | KF973475 |
| ARC-73-12 | Puerto Rico | V-African/American | 2012 | KF444913 |
| D1/Mexico/Ixtaczoquitlan/17/2007 | Mexico | V-African/American | 2007 | HM171564 |
| ThD1_0673_80 | Thailand | V-African/American | 1980 | AY732474 |
| 715393 | India | V-African/American | 1971 | JF297579 |
| IBH 28328 | Nigeria | V-African/American | 1968 | AF425625 |
| DENV-2 | ||||
| SG(EHI)D2/18944Y13 | Singapore | Cosmopolitan | 2013 | KR779784 |
| D2/IDN/Lombok_087/2012 | Bali, Indonesia | Cosmopolitan | 2012 | KM216718 |
| D2/IDN/Bali_103/2012 | Bali, Indonesia | Cosmopolitan | 2012 | KM216731 |
| D2/THA/086/2012 | Thailand | Cosmopolitan | 2012 | KM216717 |
| D2/TLS/Timor_078/2012 | East Timor | Cosmopolitan | 2012 | KM216712 |
| D2/IDN/Bali_108/2012 | Indonesia | Cosmopolitan | 2012 | KM216736 |
| 12/GZ/12851 | China | Cosmopolitan | 2012 | KF060919 |
| D2/IDN/Bali_075/2011 | Bali, Indonesia | Cosmopolitan | 2011 | KM216709 |
| D2/IN/RGCB921/2011 | India | Cosmopolitan | 2011 | KF364514 |
| 10/GZ/11864 | China | Cosmopolitan | 2010 | JN009092 |
| Philippines 2010b | Philippines | Cosmopolitan | 2010 | JN568265 |
| D2/IDN/Jakarta_060/2010 | Indonesia | Cosmopolitan | 2010 | KM216708 |
| SG(EHI)D2/63481Y10 | Singapore | Cosmopolitan | 2010 | JN030340 |
| DENV-2/VN/BID-V735/2006 | Viet Nam | Cosmopolitan | 2006 | EU482672 |
| D2/SG/05K3330DK1/2005 | Singapore | Cosmopolitan | 2005 | EU081178 |
| GWL177 INDI-01 | India | Cosmopolitan | 2001 | DQ448236 |
| 2784-DF-11/18/2002 | Taiwan | Cosmopolitan | 2002 | DQ645556 |
| MD922 | Viet Nam | Asian II | 2003 | GU434156 |
| 40247 | Brazil | Asian II | 1990 | L10041 |
| ThD2_0284_90 | Thailand | Asian II | 1990 | DQ181801 |
| PR/DB189/2013 | Puerto Rico | American/Asian | 2013 | KM279409 |
| DR59/01 | Dominican Republic | American/Asian | 2001 | AB122022 |
| Cuba115/97 | Cuba | American/Asian | 1997 | AY702050 |
| IQT-1950 | Perù | American | 1995 | DQ917242 |
| D2/TO/UH04/1974 | Tonga | American | 1974 | HM582117 |
| Laos 2010 | Laos | Asian I | 2010 | JN568244 |
| D2/Myanmar/1007aTw | Myanmar | Asian I | 2010 | JF968026 |
| D2/LAO/043/2010 | Laos | Asian I | 2010 | KM216697 |
| D2/Laos/1007aTw | Laos | Asian I | 2010 | JF968021 |
| DENV-2/KH/BID-V2062/2007 | Cambodia | Asian I | 2007 | GQ868624 |
| Myan0207a/Tw | Myanmar | Asian I | 2002 | DQ518651 |
| DENV-2/TH/BID-V2311/2001 | Thailand | Asian I | 2001 | FJ744725 |
| GD08/98 | China | Asian I | 1998 | FJ196851 |
| ThD2_0168_79 | Bangkok, Thailand | Asian I | 1979 | DQ181805 |
| DAK Ar 2022 | Burkina Faso | sylvatic | 1980 | DQ917247 |
| DAK Ar 2039 | Burkina Faso | sylvatic | 1980 | DQ917246 |
| DENV-3 | ||||
| D3/IDN/Bali_007/2010 | Indonesia | I | 2010 | KM216737 |
| D3/Malaysia/1012bTw | Malaysia | I | 2010 | JF968112 |
| H87 | Philippines | I | 1987 | M93130 |
| 80-2 | China | I | 1980 | AF317645 |
| D3-73NIID | Japan | I | 1973 | AB111085 |
| LN2632 | Malaysia | II | 1999 | AF147459 |
| D89-273 | Thailand | II | 1989 | AY145715 |
| PaH881/88 | Thailand | II | 1988 | AF349753 |
| ThD3_0046_83 | Thailand | II | 1983 | AY676358 |
| ThD3_0040_80 | Thailand | II | 1980 | AY676359 |
| ThD3_0033_74 | Thailand | II | 1974 | AY676360 |
| D3BR/ST14/04 | Brazil | III | 2004 | DQ118882 |
| - | Singapore | III | 2004 | AY662691 |
| D3PY/AS12/02 | Paraguay | III | 2002 | DQ118884 |
| BR74886/02 | Brazil | III | 2002 | AY679147 |
| Cuba_167_2001 | Cuba | III | 2001 | KT726341 |
| Cuba580/01 | Cuba | III | 2001 | AY702030 |
| D3/H/IMTSSA-SRI/2000/1266 | Sri Lanka | III | 2000 | AY099336 |
| 00-28-1HuNIID | Cambodia | III | 2000 | AB111081 |
| 6883/YUCATAN-MX/97 | Yucatan | III | 1997 | DQ341204 |
| 1339 | Puerto Rico | IV | 1997 | AY146761 |
| Human, Tahiti 1965 | Tahiti | IV | 1965 | L11439 |
| DENV-4 | ||||
| D4/RL196/Myanmar/2013 | Myanmar | I | 2013 | KJ470765 |
| D4/Thailand/0702aTw | Thailand | I | 2007 | EU448454 |
| D4/Cambodia/0509aTw | Cambodia | I | 2005 | EU448455 |
| ThD4_0485_01 | Thailand | I | 2001 | AY618992 |
| ThD4_1142_98 | Thailand | I | 1998 | AY618980 |
| ThD4_0348_91 | Bangkok, Thailand | I | 1991 | AY618990 |
| SPH317947 | Brazil | II | 2011 | JN092553 |
| DENV-4/VE/BID-V1156/2007 | Venezuela | II | 2007 | GQ868645 |
| DENV-4/CO/BID-V3412/2005 | Colombia | II | 2005 | CQ868585 |
| D4MY02-26658 | Malaysia | II | 2002 | FN429922 |
| 8976/95 | Singapore | II | 1995 | AY762085 |
| D4/PR/M35/1985 | Puerto Rico | II | 1985 | GU318316 |
| ThD4_0476_97 | Bangkok, Thailand | III | 1997 | AY618988 |
| ThD4_0017_97 | Bangkok, Thailand | III | 1997 | AY618989 |
| P75-514 | Malaysia | IV | 1975 | AF231723 |
| P73-1120 | Malaysia | IV | 1973 | AF231724 |
| CHIKV | ||||
| 10Mdy105 | Myanmar | East Central South African | 2010 | KF590567 |
| GD139 | China | East Central South African | 2010 | HQ846358 |
| TN06310 | India | East Central South African | 2010 | HM159388 |
| CU-Chik_OBF | Thailand | East Central South African | 2009 | GU908223 |
| 0901aTw | Malaysia | East Central South African | 2009 | FJ807895 |
| 0812bTw | Malaysia | East Central South African | 2008 | FJ807893 |
| CU-Chik10 | Thailand | East Central South African | 2008 | GU301780 |
| FD080178 | China | East Central South African | 2008 | GU199352 |
| 0810aTw | Bangladesh | East Central South African | 2008 | FJ807898 |
| SGEHICHT077808 | Singapore | East Central South African | 2008 | FJ445484 |
| ITA07-RA1 | Italy | East Central South African | 2007 | EU244823 |
| DRDE-07 | India | East Central South African | 2007 | EU372006 |
| LR2006_OPY1 | Reunion | East Central South African | 2006 | DQ443544 |
| 0611aTw | Singapore | East Central South African | 2006 | FJ807896 |
| SL11131 | Sri Lanka | East Central South African | 2006 | AB455493 |
| 06-027 | Reunion | East Central South African | 2005 | AM258993 |
| UgAg4155 | Uganda | East Central South African | 1982 | HM045812 |
| Vereeniging | South Africa | East Central South African | 1956 | HM045792 |
| S27-African prototype | Tanzania | East Central South African | 1953 | AF369024 |
| Ross low-psg | Tanzania | East Central South African | 1953 | HM045811 |
| TR206/H804187 | Brazil | Asian | 2014 | KP164572 |
| 0811aTw | Indonesia | Asian | 2008 | FJ807891 |
| 2008900245-BDG E1 | Indonesia | Asian | 2008 | KC879577 |
| MY021IMR/06/BP | Malaysia | Asian | 2006 | EU703762 |
| PhH15483 | Philippines | Asian | 1985 | HM045790 |
| Gibbs 63-263 | India | Asian | 1963 | HM045813 |
| TH35 | Thailand | Asian | 1958 | HM045810 |
| HD 180760 | Senegal | West African | 2005 | HM045817 |
| 37997 | Senegal | West African | 1983 | AY726732 |
Fig. 1.
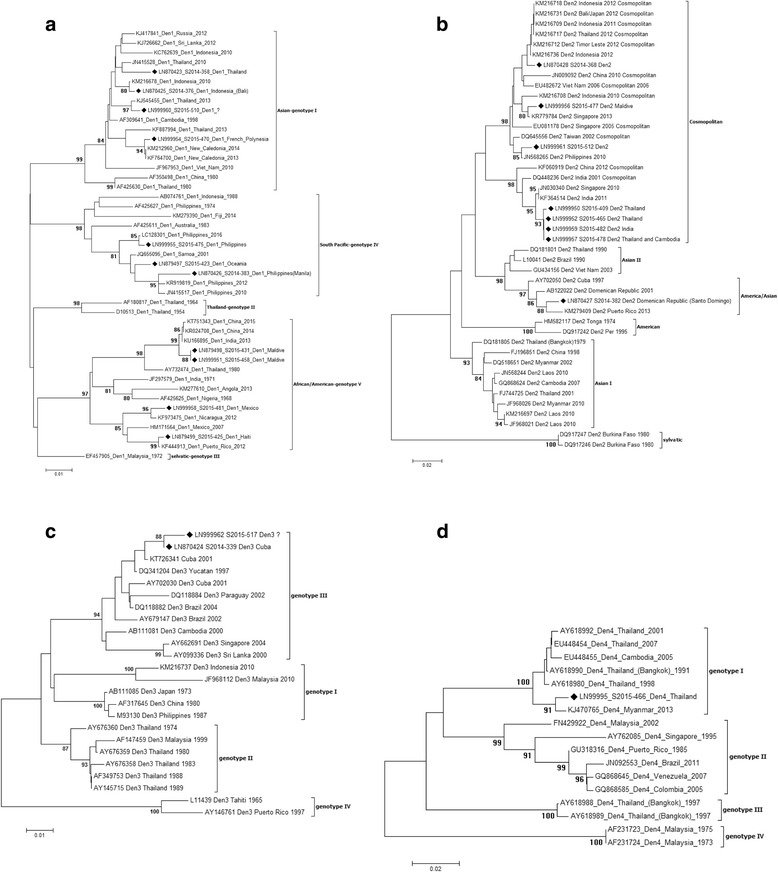
Neighbour-Joining phlylogenetic analysis of sequences obtained from DENV positive samples using Tamura-Nei model with 1000 bootstrap reiterations. For each sequence, GenBank accession number/viral genotype/country of origin of the infection/year of the infection are reported. Sequences characterized in this study are indicated by a black square. The bars indicate the percentage of diversity. Bootstrap values over 80% obtained from 1000 replicate trees are shown for key nodes. a: DENV-1 genotypes; b: DENV-2 genotypes; c: DENV-3 genotypes; d: DENV-4 genotypes
Fig. 2.
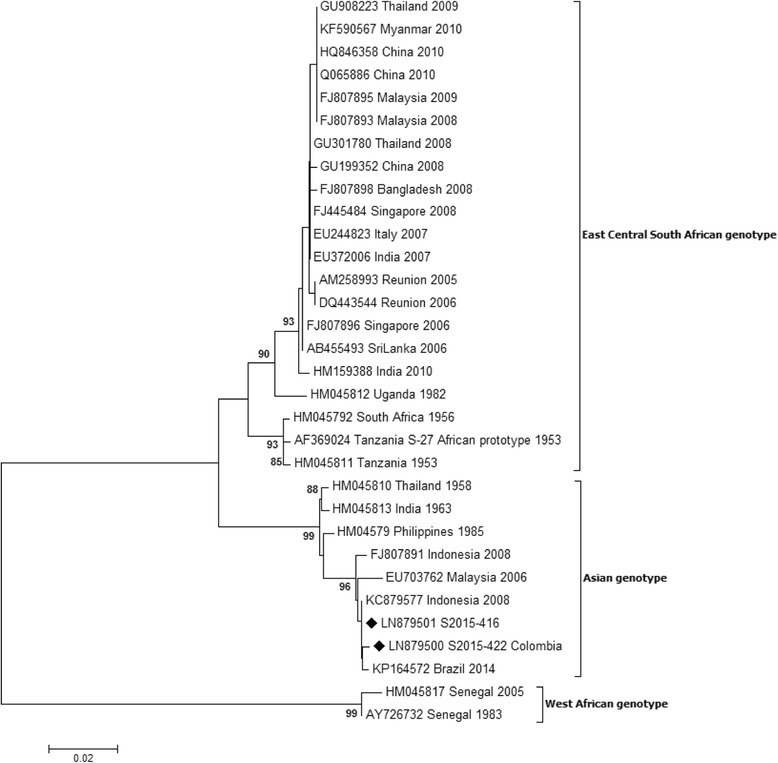
Neighbour-Joining phlylogenetic analysis of sequences obtained from CHIKV positive samples using Tamura-Nei model with 1000 bootstrap reiterations. For each sequence, GenBank accession number/viral genotype/country of origin of the infection/year of the infection are reported. Sequences characterized in this study are indicated by a black square. The bars indicate the percentage of diversity. Bootstrap values over 80% obtained from 1000 replicate trees are shown for key nodes
Discussion
In this study, we present the results of laboratory diagnosis of imported Arbovirus infections in Italy, in the period from July 2014 to October 2015. As it is well known, dengue is endemic throughout the tropics and subtropics, and its global prevalence has grown dramatically in recent years. Indeed, we found that DENV infection was the most frequently detected imported arboviral infection among our patients. Moreover, all four known DENV genotypes were detected. An increase in imported CHIKV cases was also observed, as already documented in Spain, mainland France, and Northern Italy, along with the first identification of ZIKV imported cases; both findings are attributable to the recent dramatic spread of both CHIKV and ZIKV in previously unaffected areas [27, 42–44]. The continuous expansion of the areas with Arbovirus circulation, together with the dispersion of Aedes mosquitoes spp., which are known or might be competent vectors [45], may increase the risk of outbreaks also in temperate climate areas. In Italy, the widespread presence of Ae. albopictus throughout the country, and the recent introduction and spread of new species [46, 47], make this risk particularly high. However, no autochthonous transmission chains have been recorded in our country in the period between July 2014 and October 2015.
The widespread circulation of CHIKV and ZIKV in areas until recently known to be endemic only for DENV represents a matter of concern for the potential risk of introduction in temperate regions, and raises significant diagnostic issues. In particular, problems related to the broad cross-reactivity of closely related viral agents, and the lack of well validated and standardized, commercially available tests (as it is for ZIKV), or the non-optimal performance of available tests, particularly when different viral agents co-circulate in the same areas, need to be addressed. To this regard, ZIKV imported cases are increasingly being reported, as a consequence of the continuous spread of the infection in south and central America [16, 48], leading to an increase in the requests for diagnosis. Criticisms in ZIKV diagnosis have been outlined recently [16], particularly for pregnant women [49], following the alert for the possible association between this infection and neonatal microcephaly [50].
From this scenario, the need for a careful evaluation of the diagnostic tools available for these infections clearly emerges. At this aim, in this work we have defined “our” criteria for case definition (Table 3) on the basis of the results of the diagnostic tests routinely used in our laboratory. However, it must be underlined that the final case definition for each patient is up to the clinician, and at this aim criteria are well defined in the National Plan for Arbovirus surveillance [30, 31] issued annually by the Italian Ministry of Health.
Molecular approaches for the diagnosis of viral infections are the most rapid as well as sensitive and specific. Moreover, sequencing and phylogenetic analysis of detected viruses can contribute to the knowledge of circulating viral strains and of the degree of their genetic variability. However, as suggested by our data, the use of molecular techniques is limited by the short duration of viremia during the course of the infection. With respect to serological diagnosis, we assessed some limitations in the sensitivity and/or specificity of the ELISA IgM kits routinely used in our laboratory, and also some degree of discrepancy with IgM results obtained by different laboratories. We have estimated approximately 14.5% of false positive and 11.0% false negative results for DENV, and approximately 9.6% of false negatives for CHIKV. Although the main purpose of our study was not a detailed analysis of the performances of different IgM detection systems, our data strengthen the need to confirm the diagnosis of cases defined as probable on the basis of IgM tests results. With respect to the PRNT, we have defined as “borderline” results those in which 50%,(i.e., less than 80%) of plaque reduction was observed. We found that b.l. PRNT results can be obtained in different situations. In most cases, they can be observed in samples collected soon after the onset of symptoms, and can be considered as an early, specific, response to the infection. In some cases, however, b. l. PRNT results can be due to infection by a closely related virus: we have observed a b.l. PRNT result for ZIKV in a DENV confirmed, DENV PRNT positive case; and, conversely, a DENV b.l. PRNT result in a ZIKV confirmed, ZIKV PRNT positive case. Indeed, even if PRNT is considered the most specific test, there can be some degree of cross-reactivity, thus b.l. PRNT results should always be considered cautiously. Finally, few cases showed b.l. PRNT results for several viruses (such as DENV, CHIKV and WNV), which probably represents a non-specific response of unknown origin, maybe due to an underlying, still undefined pathology.
Conclusion
DENV infection was the most frequently diagnosed cause of illness among travelers, and all four genotypes were detected. An increase in imported CHIKV cases and the first imported ZIKV cases were detected. Major diagnostic issues highlighted in our study are sensitivity limitations of molecular tests, and the importance of PRNT to confirm serological results for the differential diagnosis of Arboviruses. Moreover, the implementation of a network of laboratories involved in surveillance activities throughout the country may greatly improve the preparedness for a rapid a proper recognition of a possible autochthonous outbreak. Finally, the continuous evaluation of laboratory findings in the context of surveillance activities can be of great importance for the development of novel diagnostics, and for field evaluation of the impact of viral diseases, also in view of vaccine development and use.
Acknowledgements
The Arbovirus Working Group: Alessia Caratelli1, Veronica Bizzotti1, Daniela Casale1, Debora Lepore1, Valentina Cecchetti1, Maria Grazia Caporali3, Licia Bordi4, Fabrizio Carletti4, Francesca Colavita4, Eleonora Lalle4, Serena Quartu4, Lisa Malincarne10, Ilaria Caracciolo11, Claudia Tiberio12, Erasmo Falco12. The authors would like to thank all health professionals and clinicians from local and regional health authorities for collaborating. Maria Grazia Ciufolini for critically revising the manuscript and fruitful discussion.
Funding
None.
Availability of data and materials
Viral Sequences obtained in this study are available in the GenBank database (accession numbers are reported in the phylogenetic trees).
Authors’ contributions
GV, CF, MER and GR designed and managed this study. CR, AB, GR are involved in the epidemiological surveillance at ISS. GR is the head of the Department of Infectious, Parasitic and Immune-Mediated Diseases of the Istituto Superiore di Sanità (ISS). EB, CF, CF, MER, GV performed laboratory investigations at ISS. MER and CA performed phylogenetic analysis. GV, CF, MER and FF prepared and analysed the data. GV, CF, MER and GR contributed in writing the manuscript. CC, MRC, LZ, AB, NZ, MRG, LCN, GV, FB, PDA, GS are all involved in Arbovirus Surveillance activities, as clinicians or as responsible of Regional Reference Laboratories. All persons of the Arbovirus Working Group, both of the ISS and of Regional Reference Laboratories, contributed in laboratory investigation, and in preparing and analyzing the data. All authors read and approved the final manuscript and contributed proofreading this manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
In this study, no experiments involving recruitment of humans, nor animals, have been performed. A retrospective, aggregate analysis has been performed on human data (such as travels, symptoms, etc) from surveillance activity and from diagnostic activity performed on human clinical samples, in agreement with a National Plan issued and revised annually by Italian Ministry of Health, for which an ethics approval is not required (Decreto legislativo 24 giugno 2003, n. 211. Attuazione della direttiva 2001/20/CE relativa all’applicazione della buona pratica clinica nell’esecuzione delle sperimentazioni cliniche di medicinali per uso clinico). No data attributable to individual patients are presented in this manuscript. Clinical samples from suspected disease cases have all been collected by hospitals from several Italian Regions, according to Italian law (Decreto legislativo 30 giugno 2003, n. 196. Codice in materia di protezione dei dati personali).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- Ae.
Aedes
- b.l.
Border line
- CHIKV
Chikungunya virus
- DENV
Dengue virus
- ISS
Istituto Superiore di Sanità
- NRLA
National Reference Laboratory for Arboviruses
- PCR
Polymerase chain reaction
- PRNT
Plaque Reduction Neutralization Test
- WHO
World Health Organization
- WNV
West Nile virus
- ZIKV
Zika virus
Contributor Information
Claudia Fortuna, Email: claudia.fortuna@iss.it.
Maria Elena Remoli, Email: mariaelena.remoli@iss.it.
Caterina Rizzo, Email: caterina.rizzo@iss.it.
Eleonora Benedetti, Email: eleonora.benedetti@iss.it.
Cristiano Fiorentini, Email: cristiano.fiorentini@iss.it.
Antonino Bella, Email: antonino.bella@iss.it.
Claudio Argentini, Email: claudio.argentini@iss.it.
Francesca Farchi, Email: francesca.farchi@iss.it.
Concetta Castilletti, Email: concetta.castilletti@inmi.it.
Maria Rosaria Capobianchi, Email: maria.capobianchi@inmi.it.
Lorenzo Zammarchi, Email: lorezammarchi@gmail.com.
Alessandro Bartoloni, Email: alessandro.bartoloni@unifi.it.
Nadia Zanchetta, Email: zanchetta.nadia@asst-fbf-sacco.it.
Maria Rita Gismondo, Email: mariarita.gismondo@unimi.it.
Luca Ceccherini Nelli, Email: luca.ceccherini.nelli@gmail.com.
Giustina Vitale, Email: giustina.vitale@unipa.it.
Franco Baldelli, Email: franco.baldelli@unipg.it.
Pierlanfranco D’Agaro, Email: pierlanfranco.dagaro@burlo.trieste.it.
Giuseppe Sodano, Email: giuseppe.sodano@ospedalideicolli.it.
Giovanni Rezza, Email: giovanni.rezza@iss.it.
Giulietta Venturi, Email: giulietta.venturi@iss.it.
the Arbovirus Working Group:
Alessia Caratelli, Veronica Bizzotti, Daniela Casale, Debora Lepore, Valentina Cecchetti, Maria Grazia Caporali, Licia Bordi, Fabrizio Carletti, Francesca Colavita, Eleonora Lalle, Serena Quartu, Lisa Malincarne, Ilaria Caracciolo, Claudia Tiberio, and Erasmo Falco
References
- 1.Suk JE, Semenza JC. From global to local: vector-borne disease in an interconnected world. Eur J Public Health. 2014;24(4):531–532. doi: 10.1093/eurpub/cku041. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization . Global strategy for dengue prevention and control 2012-2020. 2012. [Google Scholar]
- 3.Cao-Lormeau VM, Musso D. Emerging arboviruses in the Pacific. Lancet. 2014;384(9954):1571–1572. doi: 10.1016/S0140-6736(14)61977-2. [DOI] [PubMed] [Google Scholar]
- 4.Weaver SC. Arrival of chikungunya virus in the new world: prospects for spread and impact on public health. PLoS Negl Trop Dis. 2014;8(6):e2921. doi: 10.1371/journal.pntd.0002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Bortel W, Dorleans F, Rosine J, Blateau A, Rousset D, Matheus S, Leparc-Goffart I, Flusin O, Prat C, Cesaire R, Najioullah F, Ardillon V, Balleydier E, Carvalho L, Lemaitre A, Noel H, Servas V, Six C, Zurbaran M, Leon L, Guinard A, van den Kerkhof J, Henry M, Fanoy E, Braks M, Reimerink J, Swaan C, Georges R, Brooks L, Freedman J, Sudre B, Zeller H. Chikungunya outbreak in the Caribbean region, December 2013 to March 2014, and the significance for Europe. Euro Surveill. 2014;19(13):20759. doi: 10.2807/1560-7917.ES2014.19.13.20759. [DOI] [PubMed] [Google Scholar]
- 6.Nunes MR, Faria NR, de Vasconcelos JM, Golding N, Kraemer MU, de Oliveira LF, Azevedo Rdo S, da Silva DE, da Silva EV, da Silva SP, Carvalho VL, Coelho GE, Cruz AC, Rodrigues SG, Vianez JL, Jr, Nunes BT, Cardoso JF, Tesh RB, Hay SI, Pybus OG, Vasconcelos PF. Emergence and potential for spread of Chikungunya virus in Brazil. BMC Med. 2015;13:102. doi: 10.1186/s12916-015-0348-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leparc-Goffart I, Nougairede A, Cassadou S, Prat C, de Lamballerie X. Chikungunya in the Americas. Lancet. 2014;383(9916):514. doi: 10.1016/S0140-6736(14)60185-9. [DOI] [PubMed] [Google Scholar]
- 8.Musso D, Cao-Lormeau VM, Gubler DJ. Zika virus: following the path of dengue and chikungunya? Lancet. 2015;386(9990):243–244. doi: 10.1016/S0140-6736(15)61273-9. [DOI] [PubMed] [Google Scholar]
- 9.Vega-Rua A, Lourenco-de-Oliveira R, Mousson L, Vazeille M, Fuchs S, Yebakima A, Gustave J, Girod R, Dusfour I, Leparc-Goffart I, Vanlandingham DL, Huang YJ, Lounibos LP, Mohamed Ali S, Nougairede A, de Lamballerie X, Failloux AB. Chikungunya virus transmission potential by local aedes mosquitoes in the americas and europe. PLoS Negl Trop Dis. 2015;9(5):e0003780. doi: 10.1371/journal.pntd.0003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayes EB. Zika virus outside Africa. Emerg Infect Dis. 2009;15(9):1347–1350. doi: 10.3201/eid1509.090442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rezza G, Nicoletti L, Angelini R, Romi R, Finarelli AC, Panning M, Cordioli P, Fortuna C, Boros S, Magurano F, Silvi G, Angelini P, Dottori M, Ciufolini MG, Majori GC, Cassone A, CHIKV study group Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet. 2007;370(9602):1840–1846. doi: 10.1016/S0140-6736(07)61779-6. [DOI] [PubMed] [Google Scholar]
- 12.Feldstein LR, Ellis EM, Rowhani-Rahbar A, Halloran EM, Ellis BR. The First Reported Outbreak of Chikungunya in the U.S. Virgin Islands, 2014-2015. Am J Trop Med Hyg. 2016;95(4):885–89. [DOI] [PMC free article] [PubMed]
- 13.Duffy MR, Chen TH, Hancock WT, Powers AM, Kool JL, Lanciotti RS, Pretrick M, Marfel M, Holzbauer S, Dubray C, Guillaumot L, Griggs A, Bel M, Lambert AJ, Laven J, Kosoy O, Panella A, Biggerstaff BJ, Fischer M, Hayes EB. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med. 2009;360(24):2536–2543. doi: 10.1056/NEJMoa0805715. [DOI] [PubMed] [Google Scholar]
- 14.Zanluca C, de Melo VC, Mosimann AL, Dos Santos GI, Dos Santos CN, Luz K. First report of autochthonous transmission of Zika virus in Brazil. Mem Inst Oswaldo Cruz. 2015;110(4):569–572. doi: 10.1590/0074-02760150192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.European Centre for Disease Prevention and Control (ECDC) Rapid Risk Assessment. Zika virus infection outbreak, Brazil and the Pacific region. 2015. [Google Scholar]
- 16.Fauci AS, Morens DM. Zika virus in the Americas - yet another arbovirus threat. N Engl J Med. 2016;374(7):601–4. doi: 10.1056/NEJMp1600297. [DOI] [PubMed] [Google Scholar]
- 17.McCarthy M. Four in Florida are infected with Zika from local mosquitoes. BMJ. 2016;354:i4235. doi: 10.1136/bmj.i4235. [DOI] [PubMed] [Google Scholar]
- 18.Oehler E, Watrin L, Larre P, Leparc-Goffart I, Lastere S, Valour F, Baudouin L, Mallet H, Musso D, Ghawche F. Zika virus infection complicated by Guillain-Barre syndrome--case report, French Polynesia, December 2013. Euro Surveill. 2014;19(9):20720. doi: 10.2807/1560-7917.ES2014.19.9.20720. [DOI] [PubMed] [Google Scholar]
- 19.World Health Organization . WHO statement on the first meeting of the International Health Regulations (2005) (IHR 2005) Emergency Committee on Zika virus and observed increase in neurological disorders and neonatal malformations. 2016. [Google Scholar]
- 20.Brasil P, Pereira JP, Jr, Raja Gabaglia C, Damasceno L, Wakimoto M, Ribeiro Nogueira RM, Carvalho de Sequeira P, Machado Siqueira A, Abreu de Carvalho LM, Cotrim da Cunha D, Calvet GA, Neves ES, Moreira ME, Rodrigues Baiao AE, Nassar de Carvalho PR, Janzen C, Valderramos SG, Cherry JD, Bispo de Filippis AM, Nielsen-Saines K. Zika Virus Infection in Pregnant Women in Rio de Janeiro - Preliminary Report. N Engl J Med. 2016;375(24):2321–2334. doi: 10.1056/NEJMoa1602412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martines RB, Bhatnagar J, Keating MK, Silva-Flannery L, Muehlenbachs A, Gary J, Goldsmith C, Hale G, Ritter J, Rollin D, Shieh WJ, Luz KG, Ramos AM, Davi HP, Kleber de Oliveria W, Lanciotti R, Lambert A, Zaki S. Notes from the field: evidence of zika virus infection in brain and placental tissues from two congenitally infected newborns and two fetal losses - Brazil, 2015. MMWR Morb Mortal Wkly Rep. 2016;65(6):159–160. doi: 10.15585/mmwr.mm6506e1. [DOI] [PubMed] [Google Scholar]
- 22.Mlakar J, Korva M, Tul N, Popovic M, Poljsak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodusek V, Vizjak A, Pizem J, Petrovec M, Avsic Zupanc T. Zika Virus Associated with Microcephaly. N Engl J Med. 2016;374(10):951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 23.Sarno M, Sacramento GA, Khouri R, do Rosario MS, Costa F, Archanjo G, Santos LA, Nery N, Jr, Vasilakis N, Ko AI, de Almeida AR. Zika Virus Infection and Stillbirths: A Case of Hydrops Fetalis, Hydranencephaly and Fetal Demise. PLoS Negl Trop Dis. 2016;10(2):e0004517. doi: 10.1371/journal.pntd.0004517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuler-Faccini L, Ribeiro EM, Feitosa IM, Horovitz DD, Cavalcanti DP, Pessoa A, Doriqui MJ, Neri JI, Neto JM, Wanderley HY, Cernach M, El-Husny AS, Pone MV, Serao CL, Sanseverino MT, Brazilian Medical Genetics Society-Zika Embryopathy Task Force Possible Association Between Zika Virus Infection and Microcephaly - Brazil, 2015. MMWR Morb Mortal Wkly Rep. 2016;65(3):59–62. doi: 10.15585/mmwr.mm6503e2. [DOI] [PubMed] [Google Scholar]
- 25.Calvet G, Aguiar RS, Melo AS, Sampaio SA, de Filippis I, Fabri A, Araujo ES, de Sequeira PC, de Mendonca MC, de Oliveira L, Tschoeke DA, Schrago CG, Thompson FL, Brasil P, Dos Santos FB, Nogueira RM, Tanuri A, de Filippis AM. Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. Lancet Infect Dis. 2016;16(6):653–60. doi: 10.1016/S1473-3099(16)00095-5. [DOI] [PubMed] [Google Scholar]
- 26.Lagi F, Zammarchi L, Strohmeyer M, Bartalesi F, Mantella A, Meli M, Blanc P, Tacconi D, Farese A, Zanelli G, Pippi F, Aquilini D, Tonziello A, Nencioni C, Benvenuti M, Moneta S, Furnari F, Ciufolini MG, Nicoletti L, Bartoloni A. Imported dengue fever in Tuscany, Italy, in the period 2006 to 2012. J Travel Med. 2014;21(5):340–343. doi: 10.1111/jtm.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zammarchi L, Tappe D, Fortuna C, Remoli ME, Gunther S, Venturi G, Bartoloni A, Schmidt-Chanasit J. Zika virus infection in a traveller returning to Europe from Brazil, March 2015. Euro Surveill. 2015;20(23):21153. doi: 10.2807/1560-7917.ES2015.20.23.21153. [DOI] [PubMed] [Google Scholar]
- 28.Napoli C, Salcuni P, Pompa MG, Declich S, Rizzo C. Estimated imported infections of Chikungunya and Dengue in Italy, 2008 to 2011. J Travel Med. 2012;19(5):294–297. doi: 10.1111/j.1708-8305.2012.00640.x. [DOI] [PubMed] [Google Scholar]
- 29.Avsic-Zupanc T. Mosquito-borne diseases--a new threat to Europe? Clin Microbiol Infect. 2013;19(8):683–684. doi: 10.1111/1469-0691.12215. [DOI] [PubMed] [Google Scholar]
- 30.Ministero della Salute, Direzione Generale della Prevenzione Sanitaria, Ufficio V, Malattie Infettive e Profilassi Internazionale ex-DGPREV: Sorveglianza dei casi umani di Chikungunya, Dengue, West Nile Disease ed altre arbovirosi e valutazione del rischio di trasmissione in Italia - 2015. Circolare 16 giugno 2015.
- 31.Ministero della Salute, Direzione Generale della Prevenzione Sanitaria, Ufficio V, Malattie Infettive e Profilassi Internazionale ex-DGPREV: Sorveglianza dei casi umani delle malattie trasmesse da vettori con particolare riferimento a Chikungunya, Dengue, Zika virus e West Nile Disease, 2014. Circolare 30 giugno 2014, n° 17674.
- 32.Baronti C, Piorkowski G, Charrel RN, Boubis L, Leparc-Goffart I, de Lamballerie X: Complete coding sequence of zika virus from a French polynesia outbreak in 2013. Genome Announc 2014, 2(3):10.1128/genomeA.00500-14. [DOI] [PMC free article] [PubMed]
- 33.Drosten C, Gottig S, Schilling S, Asper M, Panning M, Schmitz H, Gunther S. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J Clin Microbiol. 2002;40(7):2323–2330. doi: 10.1128/JCM.40.7.2323-2330.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, Duffy MR. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis. 2008;14(8):1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yenchitsomanus PT, Sricharoen P, Jaruthasana I, Pattanakitsakul SN, Nitayaphan S, Mongkolsapaya J, Malasit P. Rapid detection and identification of dengue viruses by polymerase chain reaction (PCR) Southeast Asian J Trop Med Public Health. 1996;27(2):228–236. [PubMed] [Google Scholar]
- 36.Edwards CJ, Welch SR, Chamberlain J, Hewson R, Tolley H, Cane PA, Lloyd G. Molecular diagnosis and analysis of Chikungunya virus. J Clin Virol. 2007;39(4):271–275. doi: 10.1016/j.jcv.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 37.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 39.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 40.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Venturi G, Zammarchi L, Fortuna C, Remoli ME, Benedetti E, Fiorentini C, Trotta M, Rizzo C, Mantella A, Rezza G, Bartoloni A: An autochthonous case of Zika due to possible sexual transmission, Florence, Italy, 2014. Euro Surveill 2016, 21(8):10.2807/1560-7917.ES.2016.21.8.30148. [DOI] [PubMed]
- 42.Paty MC, Six C, Charlet F, Heuze G, Cochet A, Wiegandt A, Chappert JL, Dejour-Salamanca D, Guinard A, Soler P, Servas V, Vivier-Darrigol M, Ledrans M, Debruyne M, Schaal O, Jeannin C, Helynck B, Leparc-Goffart I, Coignard B. Large number of imported chikungunya cases in mainland France, 2014: a challenge for surveillance and response. Euro Surveill. 2014;19(28):20856. doi: 10.2807/1560-7917.ES2014.19.28.20856. [DOI] [PubMed] [Google Scholar]
- 43.Rossini G, Gaibani P, Vocale C, Finarelli AC, Landini MP. Increased number of cases of Chikungunya virus (CHIKV) infection imported from the Caribbean and Central America to northern Italy, 2014. Epidemiol Infect. 2016;144(9):1912–6. [DOI] [PMC free article] [PubMed]
- 44.Requena-Mendez A, Garcia C, Aldasoro E, Vicente JA, Martinez MJ, Perez-Molina JA, Calvo-Cano A, Franco L, Parron I, Molina A, Ruiz M, Alvarez J, Sanchez-Seco MP, Gascon J. Cases of chikungunya virus infection in travellers returning to Spain from Haiti or Dominican Republic, April-June 2014. Euro Surveill. 2014;19(28):20853. doi: 10.2807/1560-7917.ES2014.19.28.20853. [DOI] [PubMed] [Google Scholar]
- 45.Di Luca M, Severini F, Toma L, Boccolini D, Romi R, Remoli ME, Sabbatucci M, Rizzo C, Venturi G, Rezza G, Fortuna C. Experimental studies of susceptibility of Italian Aedes albopictus to Zika virus. Euro Surveill. 2016;21(18):10.2807/1560-7917.ES.2016.21.18.30223. [DOI] [PubMed]
- 46.Capelli G, Drago A, Martini S, Montarsi F, Soppelsa M, Delai N, Ravagnan S, Mazzon L, Schaffner F, Mathis A, Di Luca M, Romi R, Russo F. First report in Italy of the exotic mosquito species Aedes (Finlaya) koreicus, a potential vector of arboviruses and filariae. Parasit Vectors. 2011;4:188. doi: 10.1186/1756-3305-4-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montarsi F, Drago A, Martini S, Calzolari M, De Filippo F, Bianchi A, Mazzucato M, Ciocchetta S, Arnoldi D, Baldacchino F, Rizzoli A, Capelli G. Current distribution of the invasive mosquito species, Aedes koreicus [Hulecoeteomyia koreica] in northern Italy. Parasit Vectors. 2015;8:614. doi: 10.1186/s13071-015-1208-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Attar N. ZIKA virus circulates in new regions. Nat Rev Microbiol. 2016;14(2):62. doi: 10.1038/nrmicro.2015.28. [DOI] [Google Scholar]
- 49.Petersen EE, Staples JE, Meaney-Delman D, Fischer M, Ellington SR, Callaghan WM, Jamieson DJ. Interim guidelines for pregnant women during a zika virus outbreak - United States, 2016. MMWR Morb Mortal Wkly Rep. 2016;65(2):30–33. doi: 10.15585/mmwr.mm6502e1. [DOI] [PubMed] [Google Scholar]
- 50.European Centre for Disease Prevention and Control (ECDC) Rapid risk assessment: Microcephaly in Brazil potentially linked to the Zika virus epidemic. 2015. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Viral Sequences obtained in this study are available in the GenBank database (accession numbers are reported in the phylogenetic trees).


