Fig. 1.
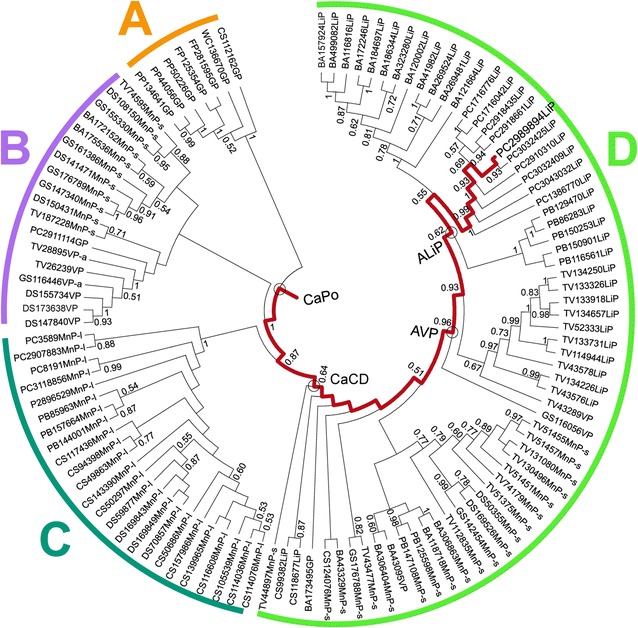
Phylogenetic tree for 113 peroxidases from ten Polyporales genomes (sequences in Additional file 2). Clusters A (GPs), B (short MnPs and VPs, plus one GP), C (long MnPs), and D (LiPs, short MnPs, and VPs, plus one GP) are shown. The path from the common ancestor to the extant LiPH8 of P. chrysosporium (JGI ID# 2989894) is in red. Also, the milestones in this evolutionary line (CaPo, CaCD, AVP, and ALiP) are marked (circles). The sequence labels start with the species code (BA, Bjerkandera adusta; CS, Ceriporiopsis subvermispora; DS, Dichomitus squalens; FP, Fomitopsis pinicola; GS, Ganoderma sp; PB, Phlebia brevispora; PC, P. chrysosporium; PP, Postia placenta; TV, Trametes versicolor; and WC, Wolfiporia cocos) followed by the JGI ID# and the peroxidase type, including GP, LiP, short MnP (MnP-s), long MnP (MnP-l), VP, and atypical VP (VP-a). Bootstrap values >0.5 are indicated on the different nodes
