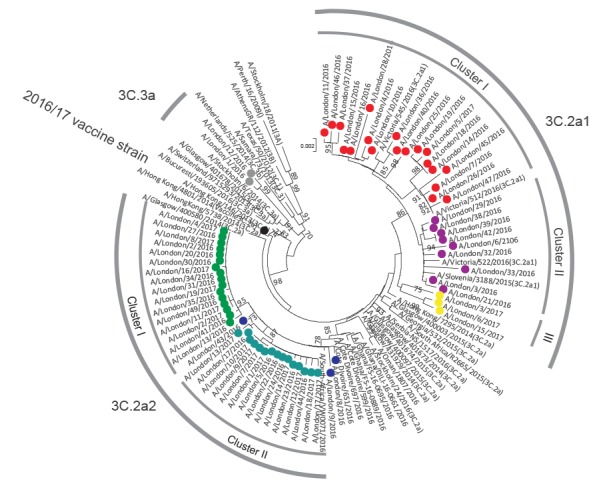
Figure 3.
Phylogenetic tree of the haemagglutinin gene sequences of virus strains recovered in this study using reference viruses for the different phylogenetic influenza A(H3N2)clades (n = 103 sequences)
Bootstrap values obtained with 100 replicates are shown on some branches of the neighbour joining tree. The scale represents the percentage of nucleotide substitutions. Sequences are annotated in the following way: influenza type/geographical area of sample collection/sample number/year. For sequences previously characterised as belonging to the subclade 3C.2a, this is indicated after the year in parentheses. London sequences retrieved in this study are indicated with coloured circles (n = 67), and the vaccine strain with a black circle.
The authors gratefully acknowledge the 36 originating and submitting laboratories who contributed sequences used in the phylogenetic analysis to the Global Initiative on Sharing All Influenza Data (GISAID; www.gisaid.org).