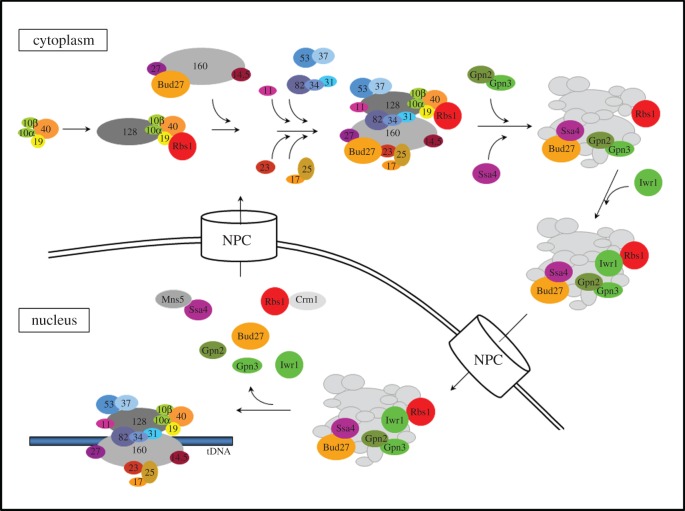
Figure 1.
Pol III biogenesis. Based on the relatively well-studied analogous process for prokaryotic RNA polymerase, it is postulated that the assembly of yeast Pol III starts with the formation of the AC19/AC40 subcomplex, probably together with the small ABC10β/ABC10α subunits, which then binds the second-largest catalytic subunit C128. The stable subcomplex C128/AC40/AC19/ABC10β/ABC10α binds the Rbs1 factor via AC40 and AC19. In a parallel step, the second major assembly intermediate is formed by the largest subunit, C160, and the ABC27 and ABC23 subunits incorporated with the help of Bud27. Pol III core is formed by joining of the two subcomplexes. Then the peripheral subunits are added as Pol III-specific subcomplexes (once the Pol III holoenzyme is assembled, Pol III subunits are presented in grey, for clarity). Gpn2, Gpn3 and Ssa4 presumably participate in later steps of Pol III biogenesis, and Iwr1 acts downstream of the GTPases and Ssa4. According to the presented model, Pol III complex is assembled in the cytoplasm prior to the nuclear import. It is also conceivable that only the core complex is formed in the cytoplasm and the peripheral subunits join it in the nucleus, as discussed in the text. Pol III is imported into the nucleus via the nuclear pore complex (NPC), probably together with the adaptors and assembly factors. The transport/assembly factors dissociate from Pol III and are exported back to the cytoplasm; Rbs1 and Ssa4 are exported, respectively, in Crm1- and Msn5-dependent manner.