Figure 2. Mapping and haplotype identification of a transcriptionally inactive paralog of the DUSP22 gene on 16p11.2.
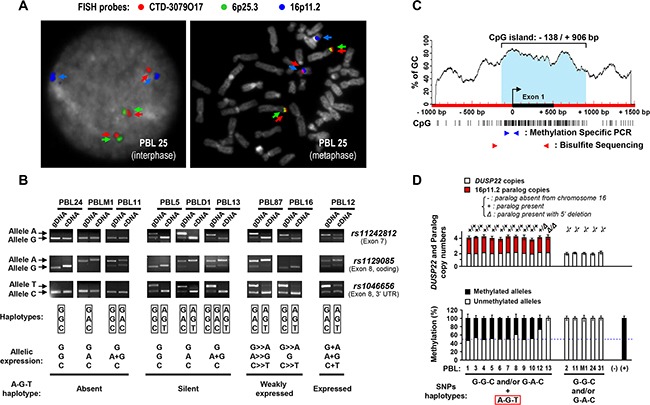
A. Metaphase and interphase FISH analysis of normal peripheral blood lymphocytes (PBL) with a 6p25.3-specific probe (RP11-164H16, Spectrum Green labeled), a probe encompassing the DUSP22 gene (CTD-3079O17, Spectrum Red labeled), and a 16p11.2-specific probe (RP11-488I20, Spectrum Gold labeled, here displayed in blue (pseudocolor) as colocalization of signals from the former probes may appear in yellow). DAPI (4′,6-diamidino-2-phenylindole dihydrochloride), here visualized in grey (pseudocolor), was used to stain nuclear DNA and chromosomes. Cases exhibiting, in addition to 6p25.3, 1 or 2 additional CTD-3079O17 probe signals, illustrate the existence of a DUSP22-related sequence subjected to copy number variations on the 16p11.2 locus (See also Supplementary S5A, B). B. Genotype (gDNA, genomic DNA) and allelic expression status (cDNA, complementary DNA) of SNPs rs11242812, rs1129085 and rs1046656 in representative cases of normal PBL, analyzed by PCR and restriction enzyme digestion (MspI, MspI and BssSI, respectively). Arrows indicate each SNP allele. SNPs genotypes and allelic expression status are summarized for each case under the electrophoresis profiles. C. Schematic representation of the CpG island (vertical bars indicate CpG sites) encompassing the 5′ region of the DUSP22 gene. The black box and curved arrow indicate exon 1 and transcription initiation site, and blue and red arrowheads the primers used for methylation specific PCR (MSP) and bisulfite genomic DNA sequencing (BGS) analyses. D. Top panel: DUSP22 and paralog copy numbers, determined by quantitative analysis of SNP rs1046656 (allele C= DUSP22; allele T= paralog), in normal PBL. KLK3 (19q13.41) was used as a control gene for normalization. Data were matched with results from3 color FISH (discriminating DUSP22 -6p25.3- from its paralog -16p11.2-, Figure 2A, Supplementary Table S6). The occurrence of deletion in the 5′ region of the paralog was evidenced by the observation of altered qPCR signal ratios between exon 1 and exon 6. Bottom panel: Quantitative MSP analysis of DUSP22/paralog 5′ CpG island in normal PBL according to the absence or presence of paralog alleles, determined as described above. No DNA (-) and in vitro methylated DNA (+) were used as negative and positive controls. Percentages of unmethylated and methylated alleles (Mean ± SEM from independent measurements) are shown.
