Figure 2. Strategy for the identification of naturally processed pentadecamer peptides.
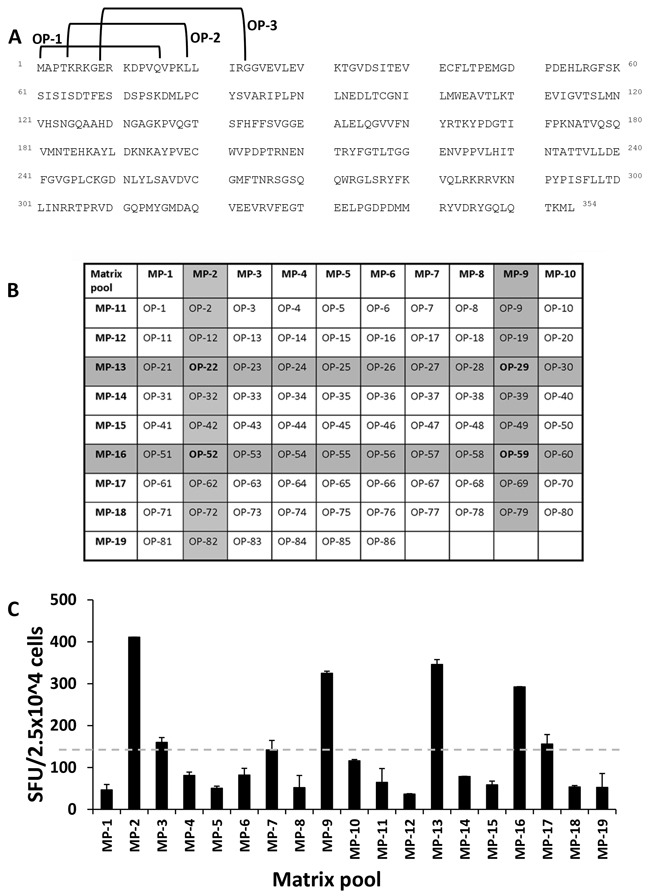
In order to identify naturally processed peptides, VP1-derived VLPs were used as a primary stimulus followed by VP1-derived overlapping peptide stimulation in MLPC assays with PBMCs from healthy donors. In the experimental setting, T2 cells served as APCs in ELISPOT assays which confirmed the HLA-A*02-restriction of identified potential peptide epitopes. A. The JCV VP1-protein (GenBank Accession No. AFH57194.1) [62] consists of 354 amino acids and was covered by 86 peptides shifted about four amino acids so that the peptides had a consecutive overlap of eleven amino acids. Exemplarily the peptides OP-1 to OP-3 are marked. OP-1 consists of amino acid 1 to 15, OP-2 of amino acid 5 to 19 and OP-3 of amino acid 9 to 23. B. These overlapping peptides were placed in a matrix grid consisting of 19 matrix pools (MP) in such a way that each peptide is uniquely contained in two intersecting pools. C. One representative example for one HLA-A*02-positive donor out of 12 healthy donors is shown. Positivity in the assay was defined as 2-fold beyond background (neg. control = 155 spots). The dashed grey line marked the background. Cells were evaluated in triplicates in each ELISPOT assay and mean and standard deviations were calculated. In this example, four MP (MP-2, MP-9, MP-13 and MP-16) were positive which corresponded to four overlapping peptides OP-22, OP-29, OP-52 and OP-59. SEB served as a positive control resulting in approximately 2,000 SFU/5 × 104 CD8+ cells.
