Figure 2. Comparison of predictive performance.
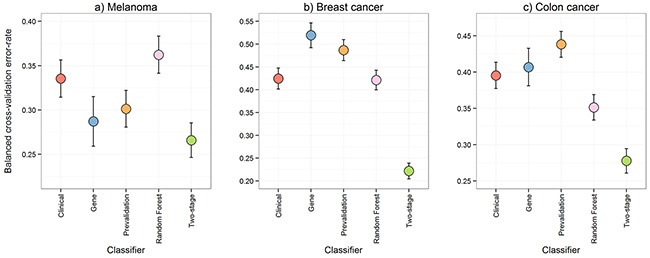
The leave-two-out cross-validation balanced error-rates from five methods performed on the melanoma, breast cancer, and colon cancer data were compared to assess predictive performance. The five methods were: 1) DLDA [16] on the gene expression data only; 2) logistic regression on the clinical variables only; 3) a pre-validation approach [9] for integrating the gene expression and clinical data; 4) Random Forests [10] on the gene expression and clinical data to capture interaction effects; and, 5) our proposed multi-step classification approach. Error bars represent the 95% confidence intervals.
