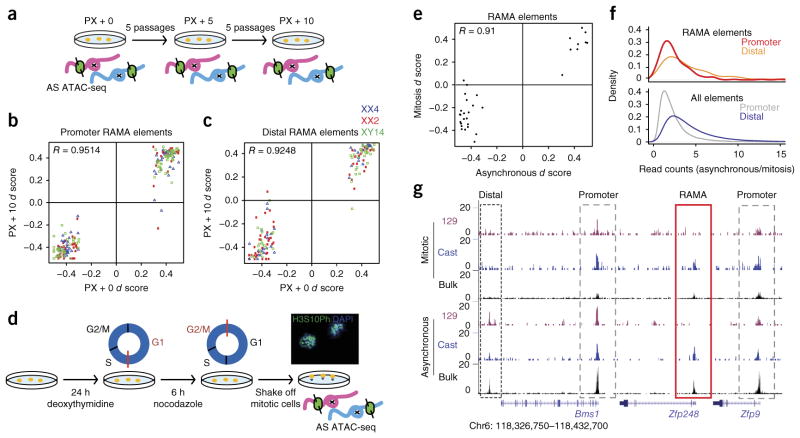
Figure 3.
RAMA elements are stable through mitosis and over many passages in the NPC state. (a) Experimental setup for allele-specific (AS) ATAC–seq across passages. Three NPC lines (XX4, XX2 and XY14) were cultured for five and ten additional passages (PX + 5 and PX + 10, respectively) after the initial ATAC–seq experiment (PX + 0). (b) ATAC–seq d scores at passage 0 versus passage 10 for promoter RAMA elements in NPC clones XX2, XX4 and XY14. (c) ATAC–seq d scores at passage 0 versus passage 10 for distal RAMA elements in NPC clones XX2, XX4 and XY14. (d) Experimental setup for mitotic ATAC–seq. NPC clone XX1 was blocked in M phase, and mitotic cells were then collected by shaking. The image shows mitotic NPCs stained with DAPI for mitotic chromosomes and antibody to H3S10ph, a marker of prometaphase cells (40× magnification). (e) ATAC–seq d scores for RAMA elements in clone XX1 in mitotic versus asynchronous cells. Monoallelic RAMA elements were included if there were ≥20 allele-informative reads under the peak in both mitotic and asynchronous ATAC–seq data. (f) Ratio of ATAC–seq read peaks in asynchronous/mitotic cells for promoter elements (<2 kb from a TSS) and distal elements (>2 kb from a TSS). RAMA elements are shown at the top, and all accessible elements are shown at the bottom. (g) Example locus showing ATAC–seq signal in mitotic and asynchronous NPCs. Allelic ATAC–seq signal is shown in magenta and blue, and non-allelic ATAC–seq signal is shown in black (bulk). Dashed black boxes indicate distal regulatory elements, dashed gray boxes indicate promoters and the red box indicates the RAMA element at the TSS of the RME gene Zfp248.