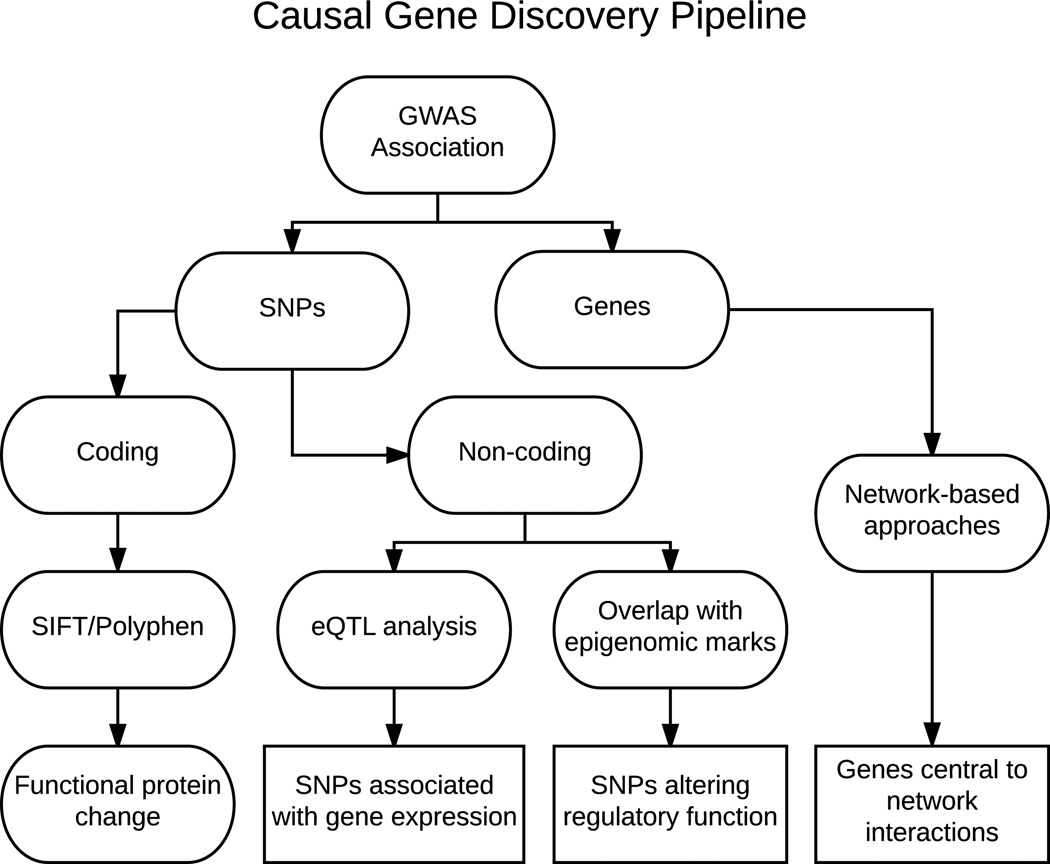
Figure 2.
Workflow for causal gene discovery. As described in Fig. 1, GWASs result in the identification of genomic loci associated with a trait or disease. Both the significantly associated SNPs and the genes within the region can be identified. If a SNP is identified within the coding region of a gene, computational techniques can be employed to determine the likelihood that the SNP causes a functional change in the protein it produces. If the SNP lies in a non-coding region, it could play a role in regulating gene expression. This hypothesis can be supported by relating the SNP to gene expression using expression quantitative trait locus (eQTL) analysis, or by SNP colocalization with regulatory epigenomic marks. The genes within the region can also be integrated with network information to identify potentially causal genes.