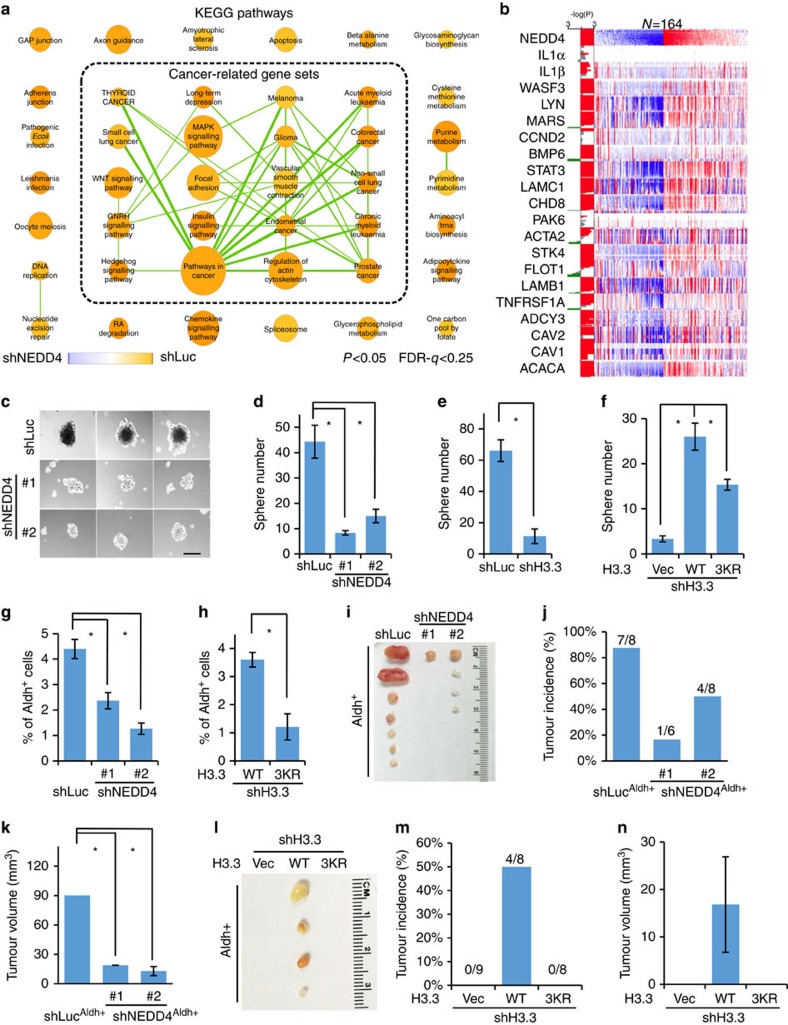
Figure 6. H3 ubiquitination is required tumour sphere forming and tumour engraftment.
(a) Cancer-related gene sets were enriched in control versus NEDD4 knockdown Hep3B cells. KEGG pathway gene sets enriched (P<0.05, q<0.25 for exploratory type of study) in control or NEDD4 knockdown were presented as orange or blue circles, respectively. Gene sets with overlapping genes were connected by green lines. The weight of the circle and line are proportional to the number of genes in gene set and overlapping genes between gene sets, respectively. See experimental procedures for details. (b) NEDD4 expression is correlated with NEDD4 target genes in TCGA hepatocarcinoma exon expression data sets. See experimental procedures for details. (c–f) NEDD4, H3.3 and H3 ubiquitination are required for in vitro tumour sphere formation. NEDD4 knockdown, H3.3 knockdown or H3.3 WT or K23/36/37R restored Hep3B cells were analysed by in vitro tumour sphere-forming assay (see experimental procedures for details). Scale bar, 300 μm in length (c). Data were presented as the mean number of three biological replicates±s.e.m. (g,h) NEDD4 and H3 ubiquitination are required for maintaining Aldh+ cell population. Control and NEDD4 knockdown or H3 WT or K23/36/37R restored Hep3B cells were stained for Aldh enzymatic activity and analysed by flow cytometry. Data were presented as the mean percentage of three biological replicates±s.e.m. See experimental procedures for details. (i–k) NEDD4 knockdown reduced in vivo tumour engraftment frequency of Du145 cells. Shown were tumour image, tumour incidence and tumour size, which was presented as the mean volume of tumours ((L × W × W)/2)±s.e.m. n=9 for each group and dead mice free of tumours are excluded. (l–n) K23/36/37R mutation reduced in vivo tumour engraftment frequency of Du145 cells. Shown were tumour image, tumour incidence and tumour size, which was presented as the mean volume of tumours ((L × W × W)/2)±s.e.m. See experimental procedures for details. All asterisks (*) represent P<0.05, using Student's T-test.