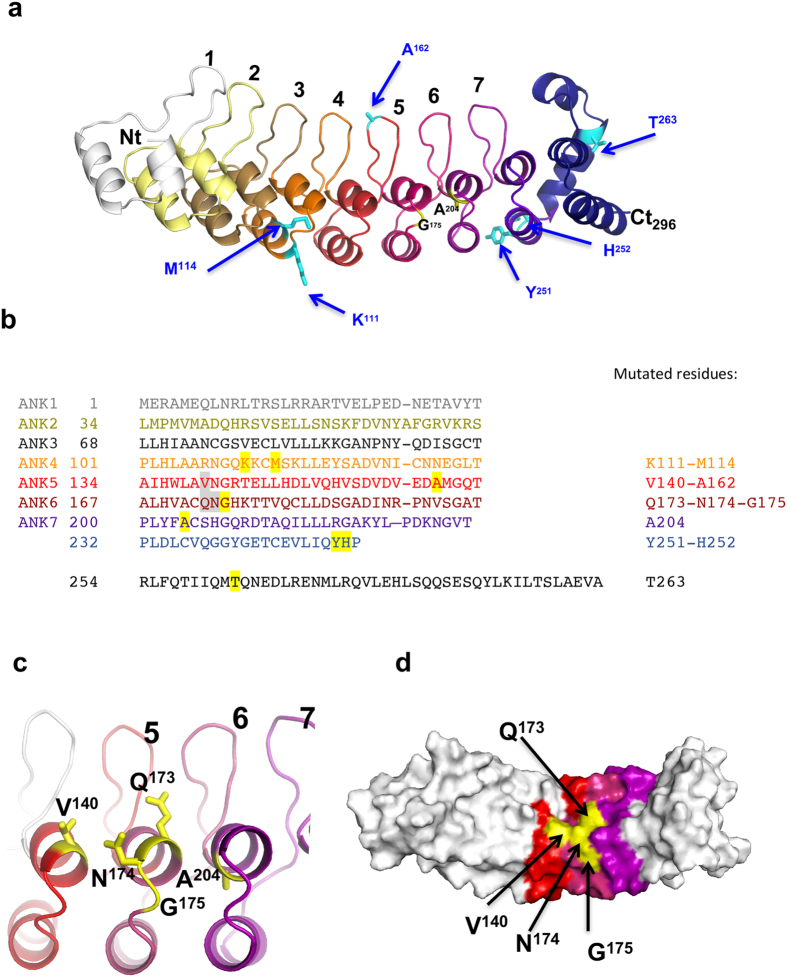
Figure 3. HACE1 mutant localization on the 3D model of HACE1 ankyrin repeats.
(a) Structural model of HACE1 calculated with the Robetta server. The model depicts the 7 ankyrin repeats at HACE1 N-terminus (residues 1–234, color-coded and labelled) followed by an incomplete ankyrin repeat (residues 235–253, dark violet) and an α-helical region (residues 254–296, dark blue). Cancer-associated missense mutations are reported on the structure. Mutation of residues in black reduced Rac1 ubiquitylation. Mutants depicted in cyan show no or slight increase in Rac1 ubiquitylation. (b) Primary sequence of HACE1 ankyrin repeats (amino acids 1–296). The 7 repeats and the incomplete 8th repeat are aligned based on the structural model. Residues 254–263 are predicted to form a helical structure that caps the last ankyrin repeat. Residues described in the text are highlighted in grey (functional epitope) and yellow (COSMIC mutations). The corresponding residues are given on the right. (c) Close-up view centered on HACE1 ankyrin repeats 5, 6 and 7 depicting surface-located amino acids V140, Q173, N174, G175. The alanine in position 204 is located on the internal part of the helix. The orientation is identical as in (a). (d) Representation of HACE1 ankyrin repeats depicting the cluster of residues controlling Rac1 ubiquitylation (yellow) at the surface of the repeats 5, 6 and 7.